# Load required packages
library(incidence2) # for uk covid daily deaths
library(EpiNow2) # to estimate time-varying reproduction number
library(epiparameter) # to access delay distributions
library(cfr) # for Ebola data (included in this package)
library(dplyr) # to format input and outputs
library(ggplot2) # to generate plots
# Set number of cores
withr::local_options(list(mc.cores = 4))
# Load Ebola data from the CFR package
data("ebola1976")
# Extract data on case onsets and format for EpiNow2
incidence_data_ebola <- ebola1976 %>%
dplyr::as_tibble() %>% # for simpler dataframe output
dplyr::select(date,cases) %>%
dplyr::rename(confirm = cases) %>%
dplyr::filter(date >= "1976-09-01")
# Preview data
incidence_data_ebola
#> # A tibble: 66 × 2
#> date confirm
#> <date> <int>
#> 1 1976-09-01 1
#> 2 1976-09-02 1
#> 3 1976-09-03 1
#> 4 1976-09-04 4
#> 5 1976-09-05 1
#> 6 1976-09-06 1
#> 7 1976-09-07 3
#> 8 1976-09-08 2
#> 9 1976-09-09 7
#> 10 1976-09-10 9
#> # ℹ 56 more rows
# Extract infection-to-death distribution (from WHO Ebola Response Team)
incubation_period_ebola_in <-
epiparameter::epiparameter_db(
disease = "ebola",
epi_name = "incubation",
single_epiparameter = TRUE
)
# Summarise distribution and type
print(incubation_period_ebola_in)
#> Disease: Ebola Virus Disease
#> Pathogen: Ebola Virus
#> Epi Parameter: incubation period
#> Study: WHO Ebola Response Team, Agua-Agum J, Ariyarajah A, Aylward B, Blake I,
#> Brennan R, Cori A, Donnelly C, Dorigatti I, Dye C, Eckmanns T, Ferguson
#> N, Formenty P, Fraser C, Garcia E, Garske T, Hinsley W, Holmes D,
#> Hugonnet S, Iyengar S, Jombart T, Krishnan R, Meijers S, Mills H,
#> Mohamed Y, Nedjati-Gilani G, Newton E, Nouvellet P, Pelletier L,
#> Perkins D, Riley S, Sagrado M, Schnitzler J, Schumacher D, Shah A, Van
#> Kerkhove M, Varsaneux O, Kannangarage N (2015). "West African Ebola
#> Epidemic after One Year — Slowing but Not Yet under Control." _The New
#> England Journal of Medicine_. doi:10.1056/NEJMc1414992
#> <https://doi.org/10.1056/NEJMc1414992>.
#> Distribution: gamma (days)
#> Parameters:
#> shape: 1.578
#> scale: 6.528
# Get parameters and format for EpiNow2 using Gamma input
incubation_ebola_params <- epiparameter::get_parameters(incubation_period_ebola_in)
# Find the upper 99.9% range by the interval
incubation_ebola_max <- round(quantile(incubation_period_ebola_in,0.999))
incubation_period_ebola <- EpiNow2::Gamma(
shape = incubation_ebola_params[["shape"]],
rate = 1/incubation_ebola_params[["scale"]],
max = incubation_ebola_max
)
# Plot delay distribution
# plot(incubation_period_ebola)
# Extract serial interval distribution distribution
# (from WHO Ebola Response Team)
serial_interval_ebola_in <-
epiparameter::epiparameter_db(
disease = "ebola",
epi_name = "serial",
single_epiparameter = TRUE
)
# Summarise distribution and type
# print(serial_interval_ebola_in)
# Discretise serial interval for input into EpiNow2
serial_int_ebola_discrete <- epiparameter::discretise(serial_interval_ebola_in)
# Find the upper 99.9% range by the interval
serial_int_ebola_discrete_max <- quantile(serial_int_ebola_discrete,0.999)
# Define parameters using LogNormal input
serial_ebola_params <- epiparameter::get_parameters(serial_int_ebola_discrete)
serial_interval_ebola <- EpiNow2::Gamma(
shape = serial_ebola_params[["shape"]],
rate = 1/serial_ebola_params[["scale"]],
max = serial_int_ebola_discrete_max
)
# Run infection estimation model
epinow_estimates <- EpiNow2::epinow(
data = incidence_data_ebola, # time series data
# assume generation time = serial interval
generation_time = generation_time_opts(serial_interval_ebola),
# delay from infection-to-death
delays = delay_opts(incubation_period_ebola),
rt = NULL,
# change default Gaussian Process priors
gp = gp_opts(alpha = Normal(0, 0.05)),
# use zero-centered prior
# instead of one centered around shifted reported cases
backcalc = backcalc_opts(prior = "none")
)
#> WARN [2025-12-08 17:30:38] epinow: Tail Effective Samples Size (ESS) is too low, indicating posterior variances and tail quantiles may be unreliable.
#> Running the chains for more iterations may help. See
#> https://mc-stan.org/misc/warnings.html#tail-ess -
# Extract infection estimates from the model output
infection_estimates <- epinow_estimates$estimates$summarised %>%
dplyr::filter(variable=="infections")
# Plot output
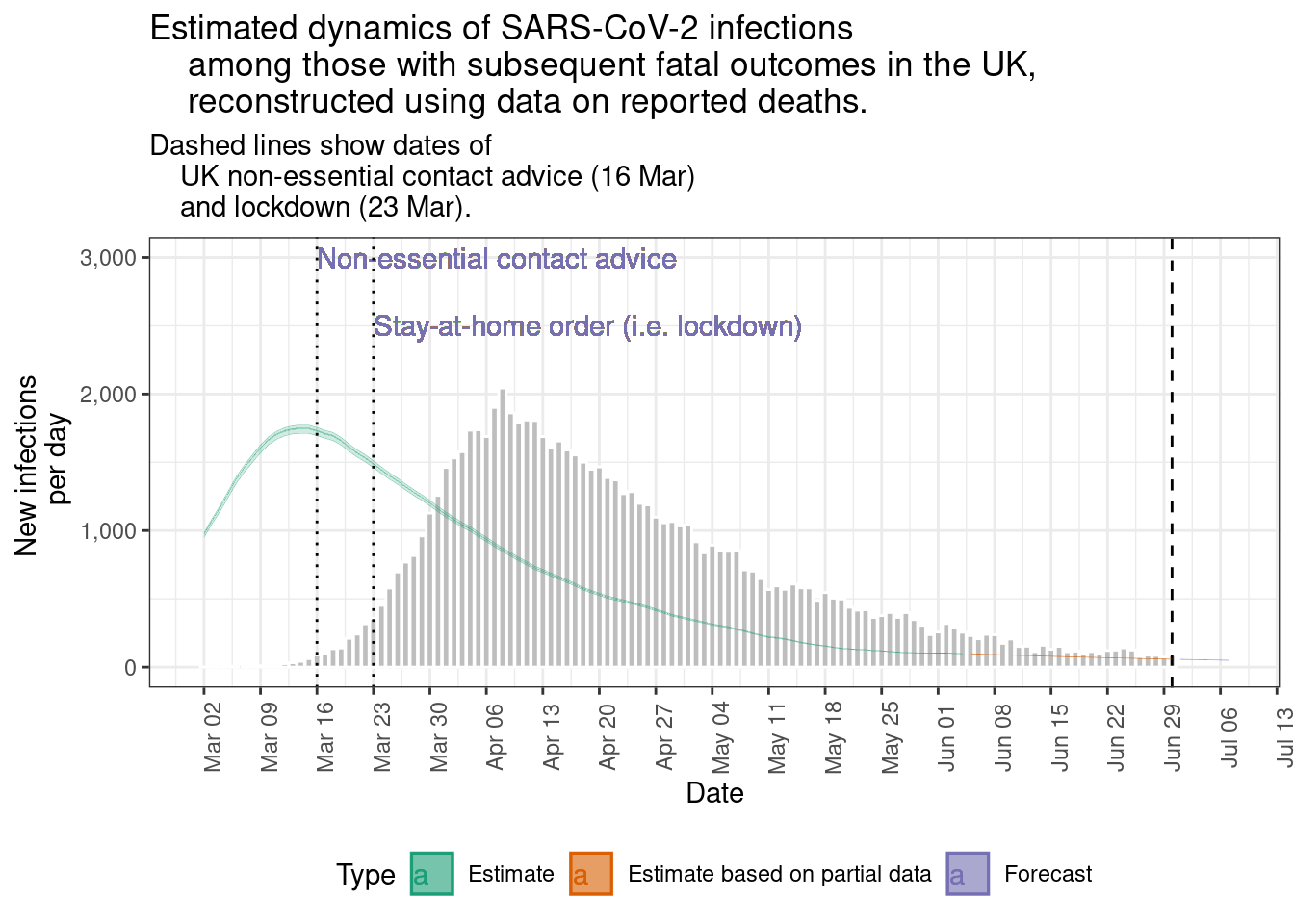
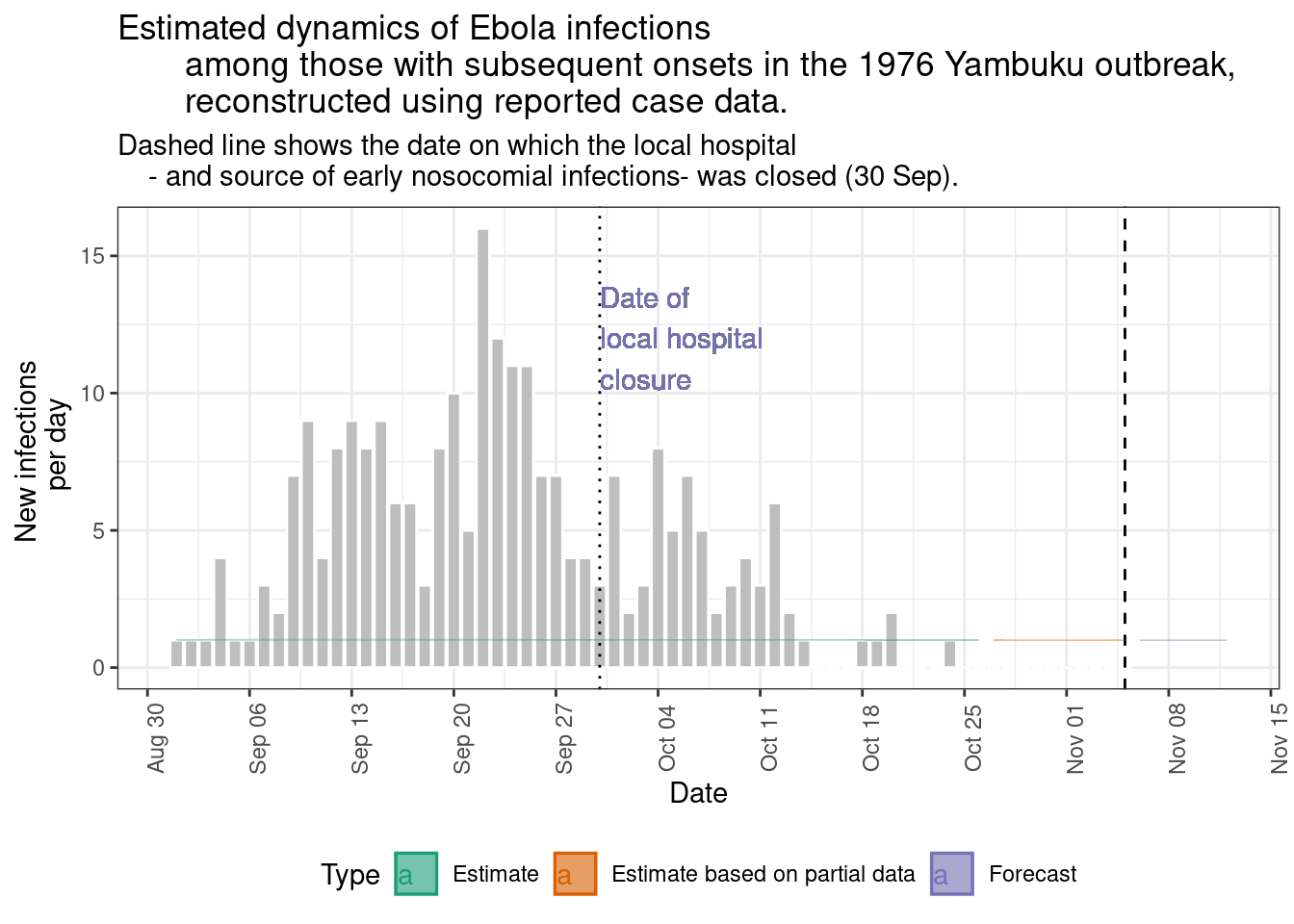
epinow_estimates$plots$infections +
geom_vline(aes(xintercept = as.Date("1976-09-30")), linetype = 3) +
geom_text(aes(x = as.Date("1976-09-30"),
y = 12,
label = "Date of\nlocal hospital\nclosure"),
hjust = 0) +
labs(
title = "Estimated dynamics of Ebola infections
among those with subsequent onsets in the 1976 Yambuku outbreak,
reconstructed using reported case data.",
subtitle = "Dashed line shows the date on which the local hospital
- and source of early nosocomial infections- was closed (30 Sep).")