# Load packages
library(epidemics)
library(socialmixr)
library(odin)
library(data.table)
library(ggplot2)
# Prepare population initial conditions
# get social contacts data
polymod <- socialmixr::polymod
contact_data <- socialmixr::contact_matrix(
polymod,
countries = "United Kingdom",
age.limits = c(0, 20, 40),
symmetric = TRUE
)
# assume arbitrary populations for each demo group
demography_vector <- contact_data$demography$population
# prepare contact matrix, divide by leading eigenvalue and rowwise by popsize
contact_matrix <- t(contact_data[["matrix"]])
contact_matrix <- (contact_matrix / max(Re(eigen(contact_matrix)$values))) /
demography_vector
# an intervention to close schools
close_schools <- epidemics::intervention(
type = "contacts",
time_begin = 200,
time_end = 260,
reduction = base::matrix(c(0.5, 0.01, 0.01))
)
# view the intervention
close_schools
#>
#> Intervention name:
#>
#> Begins at:
#> [1] 200
#>
#> Ends at:
#> [1] 260
#>
#> Reduction:
#> Interv. 1
#> Demo. grp. 1 0.50
#> Demo. grp. 2 0.01
#> Demo. grp. 3 0.01
# initial conditions defined as proportions and converted to absolute values
initial_conditions <- base::matrix(
c(S = 1 - 1e-6, E = 0, I = 1e-6, R = 0),
nrow = length(demography_vector),
ncol = 4L, byrow = TRUE
) * demography_vector
# Define an epidemic SEIR model in {odin}
seir <- odin::odin({
# NOTE: crude time-dependence of transmission rate
# similar to a `<rate_intervention>`
beta_reduce[] <- if (t > intervention_interval[1] && t < intervention_interval[2]) (1.0 - reduction[i]) else 1
# number of age groups taken from contact matrix passed by user
n <- user()
# FOI is contacts * infectious * transmission rate
# returns a matrix, must be converted to a vector/1D array
lambda_prod[, ] <- C[i, j] * I[j] * beta * beta_reduce[j]* beta_reduce[i]
#lambda_prod2[, ] <- if (i != j) lambda_prod[i, j] * beta_reduce[i] else lambda_prod[i, j] # if want to keep diagonal the same
lambda[] <- sum(lambda_prod[i, ])
## Derivatives
deriv(S[]) <- -S[i] * lambda[i]
deriv(E[]) <- (S[i] * lambda[i]) - (nu * E[i])
deriv(I[]) <- (nu * E[i]) - (gamma * I[i])
deriv(R[]) <- gamma * I[i]
## Initial conditions: passed by user
initial(S[]) <- init_S[i]
initial(E[]) <- init_E[i]
initial(I[]) <- init_I[i]
initial(R[]) <- init_R[i]
## parameters
beta <- user()
nu <- 1 / 3
gamma <- 1 / 7
C[, ] <- user() # user defined contact matrix
init_S[] <- user()
init_E[] <- user()
init_I[] <- user()
init_R[] <- user()
reduction[] <- user()
intervention_interval[] <- user()
# dimensions - all rely on contact matrix
dim(lambda_prod) <- c(n, n)
#dim(lambda_prod2) <- c(n, n)
dim(lambda) <- n
dim(S) <- n
dim(E) <- n
dim(I) <- n
dim(R) <- n
dim(init_S) <- n
dim(init_E) <- n
dim(init_I) <- n
dim(init_R) <- n
dim(reduction) <- n
dim(beta_reduce) <- n
dim(C) <- c(n, n)
dim(intervention_interval) <- 2
})
# Initialise model and run over time 0 - 600
mod <- seir$new(
beta = 1.5 / 7,
reduction = close_schools$reduction[,],
intervention_interval = c(close_schools$time_begin, close_schools$time_end),
C = contact_matrix, n = nrow(contact_matrix),
init_S = initial_conditions[, 1],
init_E = initial_conditions[, 2],
init_I = initial_conditions[, 3],
init_R = initial_conditions[, 4]
)
t <- seq(0, 600)
y <- mod$run(t)
# convert to data.table and plot infectious in each age class
y <- as.data.table(y)
y <- melt(y, id.vars = c("t"))
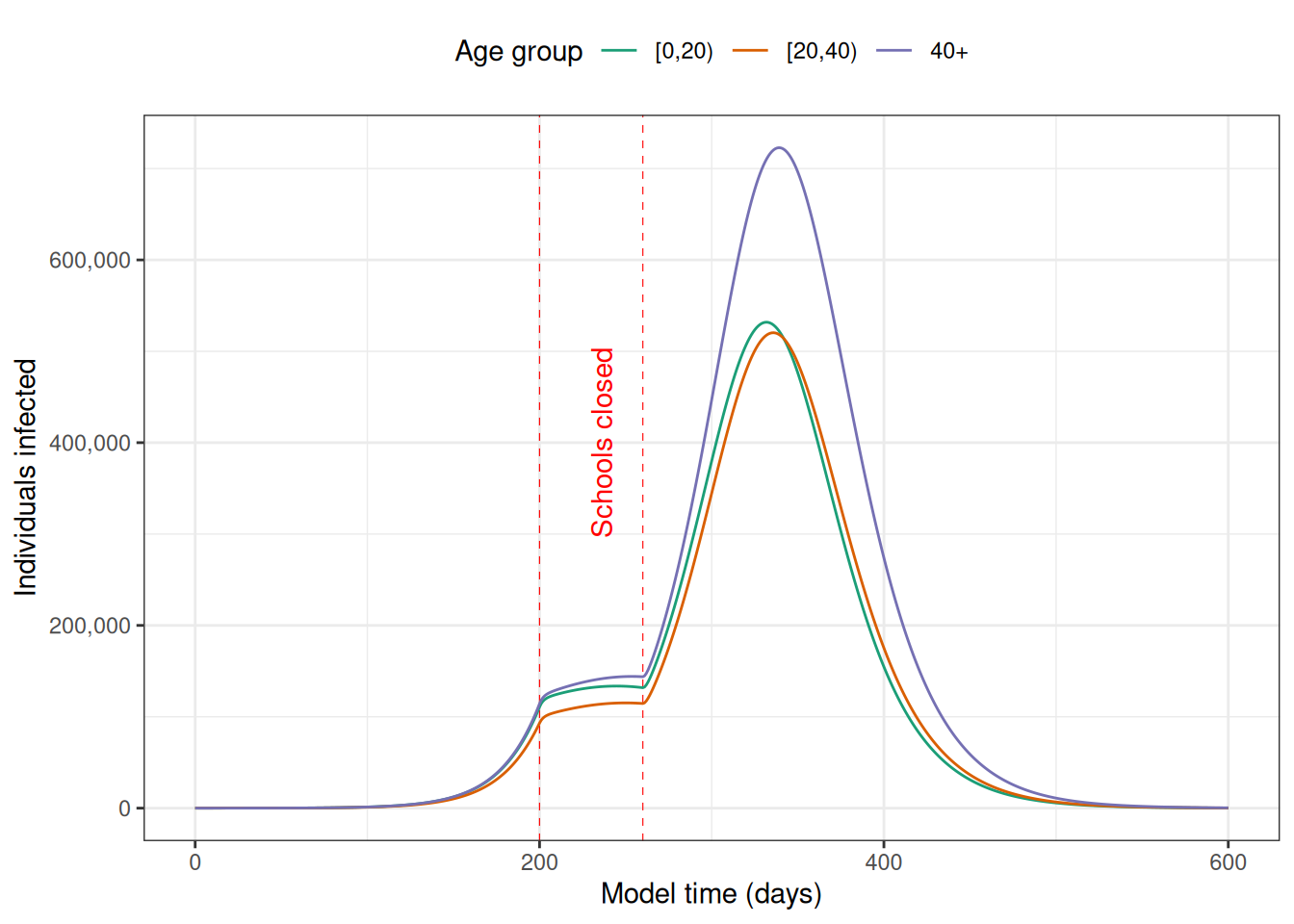
ggplot(y[variable %like% "I"]) +
geom_vline(
xintercept = c(close_schools[["time_begin"]], close_schools[["time_end"]]),
colour = "red",
linetype = "dashed",
linewidth = 0.2
) +
annotate(
geom = "text",
label = "Schools closed",
colour = "red",
x = 230, y = 400e3,
angle = 90,
vjust = "outward"
) +
geom_line(
aes(t, value, col = variable)
) +
scale_colour_brewer(
palette = "Dark2",
labels = rownames(contact_matrix),
name = "Age group"
) +
scale_y_continuous(
labels = scales::comma,
name = "Individuals infected"
) +
labs(
x = "Model time (days)"
) +
theme_bw() +
theme(
legend.position = "top"
)