This function constructs an S3 object of the class
vaccineff_data that contains all the relevant information for the study.
to estimate the effectiveness.
Usage
make_vaccineff_data(
data_set,
outcome_date_col,
censoring_date_col = NULL,
vacc_date_col,
vacc_name_col = NULL,
vaccinated_status = "v",
unvaccinated_status = "u",
immunization_delay = 0,
end_cohort,
match = FALSE,
exact = NULL,
nearest = NULL,
take_first = FALSE,
t0_follow_up = NULL
)Arguments
- data_set
data.framewith cohort information.- outcome_date_col
Name of the column that contains the outcome dates.
- censoring_date_col
Name of the column that contains the censoring date. NULL by default.
- vacc_date_col
Name of the column(s) that contain the vaccine dates.
- vacc_name_col
Name of the column(s) that contain custom vaccine names for the vaccines (e.g. brand name, type of vaccine). If provided, must be of the same length as
vacc_date_col.- vaccinated_status
Status assigned to the vaccinated population. Default is
v.- unvaccinated_status
Status assigned to the unvaccinated population. Default is
u.- immunization_delay
Characteristic time in days before the patient is considered immune. Default is 0.
- end_cohort
End date of the study.
- match
TRUE: cohort matching is performed. Default isFALSE- exact
Name(s) of column(s) for
exactmatching. Default isNULL.- nearest
Named vector with name(s) of column(s) for
nearestmatching and caliper(s) for each variable (e.g.,nearest = c("characteristic1" = n1, "characteristic2" = n2), wheren1andn2are the calipers). Default isNULL.- take_first
FALSE: takes the latest vaccine date.TRUE: takes the earliest vaccine date.- t0_follow_up
Column with the initial dates of the follow-up period. This column is only used if
match = FALSE. If not provided, the follow-up period starts atstart_cohort. Default is NULL.
Value
An S3 object of class vaccineff_data with all the information and
characteristics of the study. data.frames are converted into an object of
class linelist to easily handle with the data.
Examples
# \donttest{
# Load example data
data("cohortdata")
# Create `vaccineff_data`
vaccineff_data <- make_vaccineff_data(data_set = cohortdata,
outcome_date_col = "death_date",
censoring_date_col = "death_other_causes",
vacc_date_col = "vaccine_date_2",
vaccinated_status = "v",
unvaccinated_status = "u",
immunization_delay = 15,
end_cohort = as.Date("2021-12-31"),
match = TRUE,
exact = c("age", "sex"),
nearest = NULL
)
# Print summary of data
summary(vaccineff_data)
#> Cohort start: 2021-03-26
#> Cohort end: 2021-12-31
#> The start date of the cohort was defined as the mininimum immunization date.
#> 65 registers were removed with outcomes before the start date.
#>
#> Nearest neighbors matching iteratively performed.
#> Number of iterations: 4
#> Balance all:
#> u v smd
#> age 63.917069 62.997438 -0.08593156
#> sex_F 0.520277 0.573474 0.10701746
#> sex_M 0.479723 0.426526 -0.10701746
#>
#> Balance matched:
#> u v smd
#> age 63.751784 63.751784 0
#> sex_F 0.521457 0.521457 0
#> sex_M 0.478543 0.478543 0
#>
#> Summary vaccination:
#> u v
#> All 10973 19905
#> Matched 10789 10789
#> Unmatched 184 9116
#>
#> // tags: outcome_date_col:death_date, censoring_date_col:death_other_causes, vacc_date_col:vaccine_date_2, immunization_date_col:immunization_date, vacc_status_col:vaccine_status
# Plot vaccine coverage
plot(vaccineff_data)
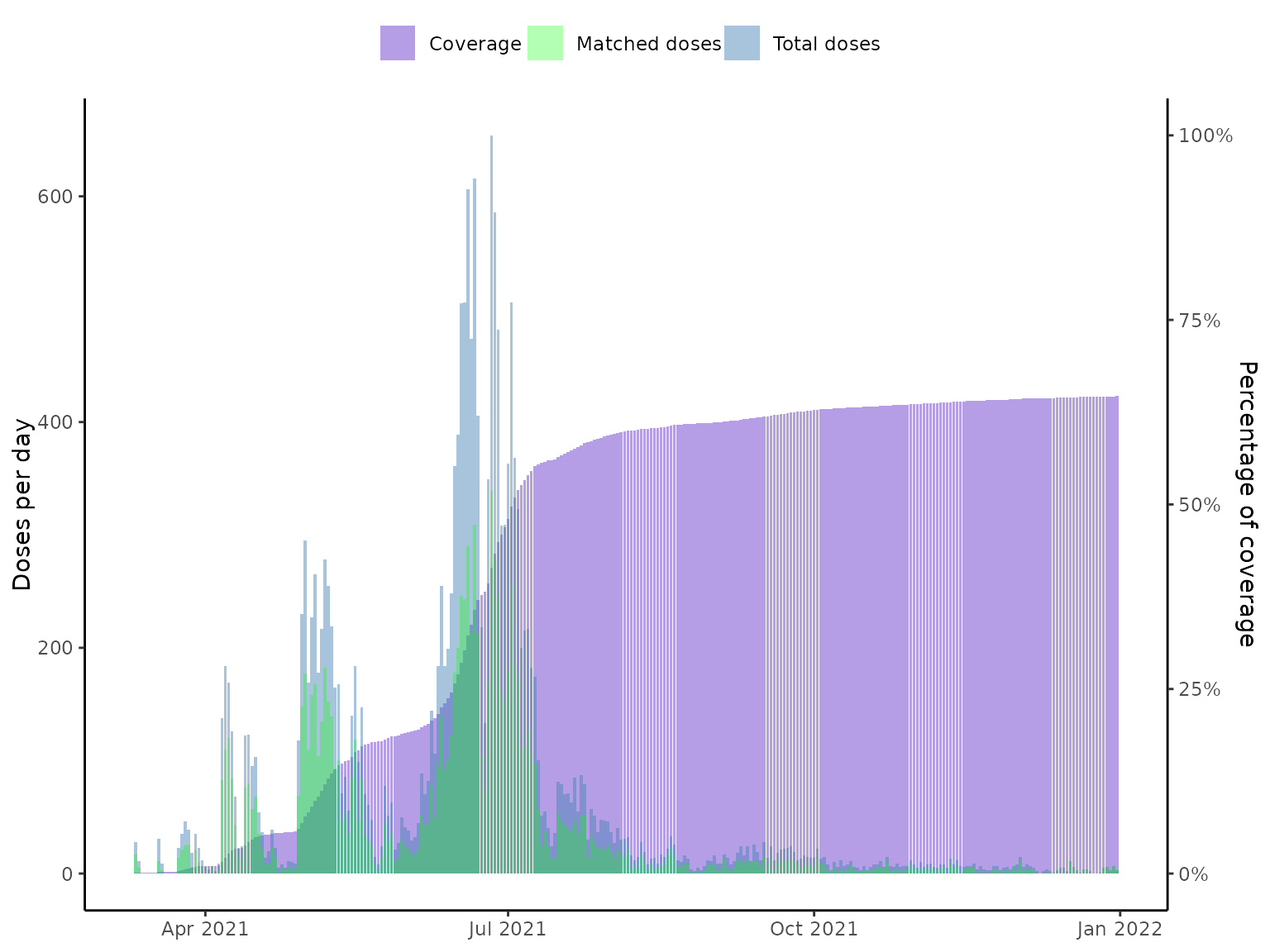 # }
# }
