Introduction to episoap: A Store of Outbreak Analytics Pipelines
Source:vignettes/episoap.Rmd
episoap.RmdThis package provides a store of curated outbreak analytics pipelines as rmarkdown reports.
Loading reports
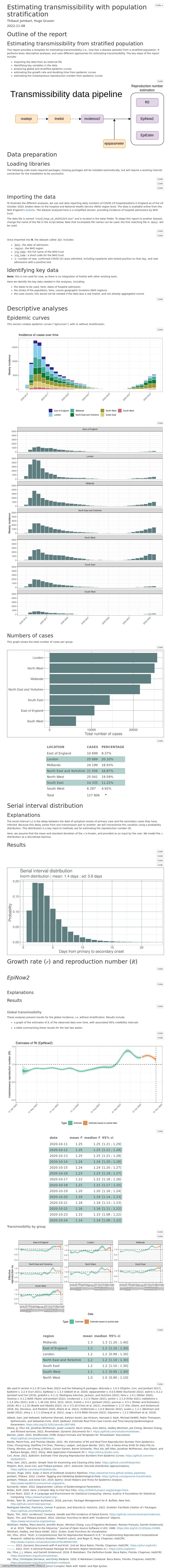
Reports are provided a rmarkdown templates. You can load them either via RStudio graphical interface
or by running:
rmarkdown::draft(
file = "myreport.Rmd",
template = "transmissibility",
package = "episoap"
)To get a list of the template reports available in this package, you can run:
episoap::list_templates()
#> [1] "transmissibility"Customising reports
# Load template in temporary directory for this vignette. You will probably
# wish to save it somewhere else.
rmarkdown::draft(
file = file.path(tempdir(), "myreport"),
template = "transmissibility",
package = "episoap",
edit = FALSE
)The reports are classic rmarkdown files. If you are familiar with rmarkdown and literate programming, you can edit any piece of the report as you see fit.
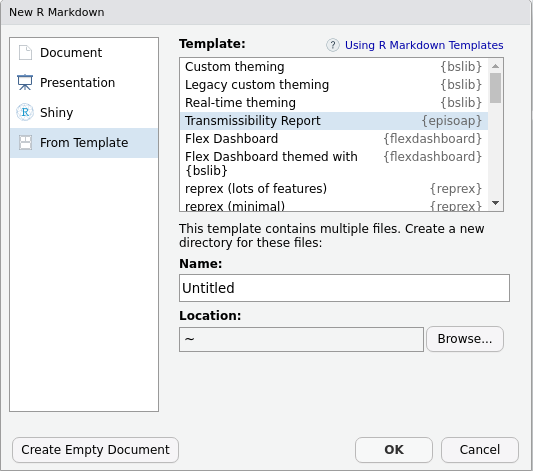
For users who want a more ready-to-use solution, reports are already
set up to provide multiple options for your analysis out of the box. For
example, in the transmissibility report, you can choose to
estimate the reproduction number in your data with different tools, such
as EpiEstim or EpiNow2. This is achieved
by changing the value of the rt_estimator parameter.
Again, this can be done with a graphical interface by using the ‘Knit with parameters’ button in RStudio:
or by running:
# Get a graphical interface to select the parameters to use in this report
rmarkdown::render(
input = file.path(tempdir(), "myreport", "myreport.Rmd"),
output_file = "custom_report.html",
params = "ask",
quiet = TRUE
)Alternatively, you can also directly the value of the various
parameters in the rmarkdown::render() call:
# Report using EpiEstim to estimate Rt
rmarkdown::render(
input = file.path(tempdir(), "myreport", "myreport.Rmd"),
output_file = "EpiEstim_report.html",
params = list(rt_estimator = "EpiEstim"),
quiet = TRUE
)
# Report using R0 to estimate Rt
rmarkdown::render(
input = file.path(tempdir(), "myreport", "myreport.Rmd"),
output_file = "EpiNow2_report.html",
params = list(rt_estimator = "EpiNow2"),
quiet = TRUE
)