This package provides a store of curated outbreak analytics pipelines as rmarkdown reports.
Target audience
The analyses are largely automated, and should be of use to any outbreak analyst. A basic R literacy will be required to adapt the report to other datasets.
Installation
You can install the development version of soap from GitHub with:
# install.packages("devtools")
devtools::install_github("epiverse-trace/episoap")Usage
Reports are provided a rmarkdown templates. You can load them either via RStudio graphical interface
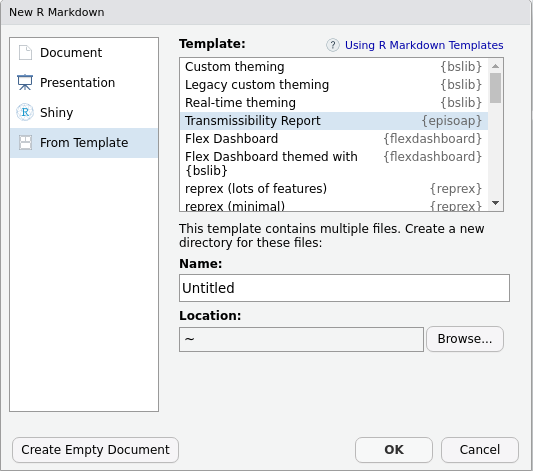
or by running:
rmarkdown::draft(file = "myreport.Rmd", template = "transmissibility", package = "episoap")To get a list of the template reports available in this package, you can run:
episoap::list_templates()
#> [1] "transmissibility"Related projects
This project has some overlap with other R packages:
-
{sitrep}from the Applied Epi organisation. While the stated goals and approaches can appear similar, episoap and{sitrep}are actually two very different projects. The{sitrep}reports are more specific (providing, e.g., reports for a specific disease although a generic template is in development), and thus more detailed. They are also more opinionated in the sense that they provide a single analysis path for each situation, based on the extensive experience of MSF. episoap on the other hand offers a more generic approach, with the emphasis on alternative paths you can take within a single analysis.
Acknowledgements
- Thanks to Sam Abbott for pointing out issues with the way EpiNow2 was used in the transmissibility pipeline (#35)
- The package logo is a derivative from a pipeline logo, provided by flaticon user “Eucalyp” for free for personal and commercial use with attribution.