## Clean and validate and plot outbreak data
# Load required R packages ------------------------------------------------
library(simulist)
library(cleanepi)
library(numberize)
library(incidence2)
library(tibble)
library(tidyr)
library(dplyr)
# Choose a seed that results in suitable and reproducible outbreak --------
set.seed(1)
# Simulate outbreak -------------------------------------------------------
line_list <- simulist::sim_linelist() %>%
# to tibble for tidier printing
tibble::as_tibble()
line_list
#> # A tibble: 158 × 13
#> id case_name case_type sex age date_onset date_reporting
#> <int> <chr> <chr> <chr> <int> <date> <date>
#> 1 1 Lolette Phillips suspected f 59 2023-01-01 2023-01-01
#> 2 2 James Jack suspected m 90 2023-01-01 2023-01-01
#> 3 3 Chen Kantha confirmed m 4 2023-01-02 2023-01-02
#> 4 5 Saleema al-Zaki probable f 29 2023-01-04 2023-01-04
#> 5 6 David Ponzio confirmed m 14 2023-01-05 2023-01-05
#> 6 7 Christopher Ward probable m 85 2023-01-06 2023-01-06
#> 7 10 Laura Ilaoa confirmed f 25 2023-01-13 2023-01-13
#> 8 11 Morgan Mason suspected f 34 2023-01-11 2023-01-11
#> 9 12 Cornelius Turner confirmed m 89 2023-01-13 2023-01-13
#> 10 14 Shaddaad el-Younes suspected m 63 2023-01-11 2023-01-11
#> # ℹ 148 more rows
#> # ℹ 6 more variables: date_admission <date>, outcome <chr>,
#> # date_outcome <date>, date_first_contact <date>, date_last_contact <date>,
#> # ct_value <dbl>
# Create messy line list data ---------------------------------------------
line_list <- simulist::messy_linelist(line_list, inconsistent_dates = TRUE)
line_list
#> # A tibble: 174 × 13
#> id case_name case_type sex age date_onset date_reporting
#> <chr> <chr> <chr> <chr> <chr> <chr> <chr>
#> 1 1 Lolette Phillips suspected <NA> 59 01 Januar… <NA>
#> 2 two James Jack suspected <NA> 90 01-01-2023 01-01-2023
#> 3 3 Chen Kantha confirued M four <NA> <NA>
#> 4 five <NA> probable <NA> twenty… 04-01-2023 04-01-2023
#> 5 6 David Ponzio confirmed myle fourte… 05 Jan 20… 05 Jan 2023
#> 6 seven Christopher Ward probable mmle eighty… 06-01-2023 06-01-2023
#> 7 10 Laura Ilaoa <NA> <NA> twenty… 13 Januar… 13 January 20…
#> 8 11 Morgan Mason suspected f <NA> 11 Jan 20… 11 Jan 2023
#> 9 12 Cornelius Turner confirmed M eighty… <NA> 13-01-2023
#> 10 fourteen Shaddaad el-Younes suspected Male 63 2023/01/11 2023/01/11
#> # ℹ 164 more rows
#> # ℹ 6 more variables: date_admission <chr>, outcome <chr>, date_outcome <chr>,
#> # date_first_contact <chr>, date_last_contact <chr>, ct_value <chr>
# Tag line list of data validation ----------------------------------------
# see what tags are available
linelist::tags_names()
#> [1] "id" "date_onset" "date_reporting" "date_admission"
#> [5] "date_discharge" "date_outcome" "date_death" "gender"
#> [9] "age" "location" "occupation" "hcw"
#> [13] "outcome"
# in this case the tags have the same name but line list columns can be
# named differently from the tag names
line_list <- linelist::make_linelist(
x = line_list,
date_onset = "date_onset",
date_admission = "date_admission",
date_outcome = "date_outcome"
)
line_list
#>
#> // linelist object
#> # A tibble: 174 × 13
#> id case_name case_type sex age date_onset date_reporting
#> <chr> <chr> <chr> <chr> <chr> <chr> <chr>
#> 1 1 Lolette Phillips suspected <NA> 59 01 Januar… <NA>
#> 2 two James Jack suspected <NA> 90 01-01-2023 01-01-2023
#> 3 3 Chen Kantha confirued M four <NA> <NA>
#> 4 five <NA> probable <NA> twenty… 04-01-2023 04-01-2023
#> 5 6 David Ponzio confirmed myle fourte… 05 Jan 20… 05 Jan 2023
#> 6 seven Christopher Ward probable mmle eighty… 06-01-2023 06-01-2023
#> 7 10 Laura Ilaoa <NA> <NA> twenty… 13 Januar… 13 January 20…
#> 8 11 Morgan Mason suspected f <NA> 11 Jan 20… 11 Jan 2023
#> 9 12 Cornelius Turner confirmed M eighty… <NA> 13-01-2023
#> 10 fourteen Shaddaad el-Younes suspected Male 63 2023/01/11 2023/01/11
#> # ℹ 164 more rows
#> # ℹ 6 more variables: date_admission <chr>, outcome <chr>, date_outcome <chr>,
#> # date_first_contact <chr>, date_last_contact <chr>, ct_value <chr>
#>
#> // tags: date_onset:date_onset, date_admission:date_admission, date_outcome:date_outcome
# line list can be validated using tags
# this will error due to the line list being messy
# linelist::validate_linelist(line_list)
# Scan line list data for issues ------------------------------------------
# see {cleanepi} website: https://epiverse-trace.github.io/cleanepi/
cleanepi::scan_data(line_list)
#> Field_names missing numeric date character logical
#> 1 id 0.0943 0.5157 0 0.4843 0
#> 2 case_name 0.1678 0.0000 0 1.0000 0
#> 3 case_type 0.0875 0.0000 0 1.0000 0
#> 4 sex 0.1678 0.0000 0 1.0000 0
#> 5 age 0.1154 0.5128 0 0.4872 0
#> 6 date_onset 0.1447 0.0000 1 0.0000 0
#> 7 date_reporting 0.2254 0.0000 1 0.0000 0
#> 8 date_admission 4.4375 0.0000 1 0.0000 0
#> 9 outcome 0.1447 0.0000 0 1.0000 0
#> 10 date_outcome 5.9600 0.0000 1 0.0000 0
#> 11 date_first_contact 0.1600 0.0000 1 0.0000 0
#> 12 date_last_contact 0.0807 0.0000 1 0.0000 0
#> 13 ct_value 1.1481 1.0000 0 0.0000 0
# Clean line list ---------------------------------------------------------
line_list$age <- numberize::numberize(line_list$age)
line_list$age
#> [1] 59 90 4 29 14 85 25 NA 89 63 74 34 29 46 24 41 72 6 61 44 71 69 69 61 64
#> [26] 61 66 66 81 81 53 45 45 42 78 35 54 68 3 44 62 62 35 90 25 46 8 NA 54 67
#> [51] 37 73 13 72 61 22 22 29 17 17 NA 90 90 23 NA 62 NA 44 57 57 63 46 66 68 12
#> [76] 51 2 53 NA 82 25 NA 14 76 78 75 76 80 80 41 NA 28 2 82 5 NA 44 76 3 15
#> [101] 19 76 90 64 NA NA 18 79 49 56 76 41 41 29 39 32 9 9 39 12 52 NA 68 57 8
#> [126] 28 2 84 52 58 29 30 71 43 40 40 NA NA 69 38 28 82 19 46 71 4 4 NA 16 53
#> [151] 14 48 89 39 NA NA 28 42 71 9 61 53 76 69 50 7 7 45 45 83 NA 88 79 82
line_list$id <- numberize::numberize(line_list$id)
line_list$id
#> [1] 1 2 3 5 6 7 10 11 12 14 18 19 22 24 27 29 30 31
#> [19] 33 34 35 NA NA 39 41 44 45 45 47 47 50 51 51 53 NA 55
#> [37] 56 58 59 60 63 63 64 65 66 71 73 77 78 80 85 NA 89 90
#> [55] 93 97 97 98 101 101 108 111 111 113 114 116 119 120 NA NA 123 125
#> [73] 126 130 131 133 138 139 142 143 145 146 147 150 153 154 155 157 157 158
#> [91] 159 163 164 166 171 172 173 NA 175 176 177 178 182 NA 189 195 196 197
#> [109] 199 200 201 202 202 205 209 NA 211 211 213 214 219 NA 224 NA 229 230
#> [127] 232 233 234 235 236 238 240 NA 245 245 250 NA NA 253 254 255 258 261
#> [145] 262 264 264 265 NA 268 277 278 279 280 282 284 285 291 292 293 296 298
#> [163] 301 302 304 307 307 310 310 311 313 316 320 326
cleanepi::check_subject_ids(line_list, target_columns = "id", range = c(1, nrow(line_list)))
#>
#> // linelist object
#> # A tibble: 174 × 13
#> id case_name case_type sex age date_onset date_reporting
#> <chr> <chr> <chr> <chr> <dbl> <chr> <chr>
#> 1 1 Lolette Phillips suspected <NA> 59 01 January 2023 <NA>
#> 2 2 James Jack suspected <NA> 90 01-01-2023 01-01-2023
#> 3 3 Chen Kantha confirued M 4 <NA> <NA>
#> 4 5 <NA> probable <NA> 29 04-01-2023 04-01-2023
#> 5 6 David Ponzio confirmed myle 14 05 Jan 2023 05 Jan 2023
#> 6 7 Christopher Ward probable mmle 85 06-01-2023 06-01-2023
#> 7 10 Laura Ilaoa <NA> <NA> 25 13 January 2023 13 January 20…
#> 8 11 Morgan Mason suspected f NA 11 Jan 2023 11 Jan 2023
#> 9 12 Cornelius Turner confirmed M 89 <NA> 13-01-2023
#> 10 14 Shaddaad el-Younes suspected Male 63 2023/01/11 2023/01/11
#> # ℹ 164 more rows
#> # ℹ 6 more variables: date_admission <chr>, outcome <chr>, date_outcome <chr>,
#> # date_first_contact <chr>, date_last_contact <chr>, ct_value <chr>
#>
#> // tags: date_onset:date_onset, date_admission:date_admission, date_outcome:date_outcome
# routine cleaning steps to tidy column names and remove duplicated rows
line_list <- line_list %>%
cleanepi::standardize_column_names() %>%
cleanepi::remove_constants() %>%
cleanepi::remove_duplicates()
date_columns <- colnames(line_list)[startsWith(colnames(line_list), "date_")]
line_list <- line_list %>%
cleanepi::standardize_dates(target_columns = date_columns)
# clean inconsistent sex using dictionary ---------------------------------
# Find inconsistencies
line_list %>% count(sex)
#>
#> // linelist object
#> # A tibble: 15 × 2
#> sex n
#> <chr> <int>
#> 1 F 22
#> 2 Female 16
#> 3 M 15
#> 4 Male 19
#> 5 f 16
#> 6 female 14
#> 7 femvle 1
#> 8 femyle 1
#> 9 m 17
#> 10 malb 1
#> 11 mald 1
#> 12 male 9
#> 13 mmle 1
#> 14 myle 2
#> 15 <NA> 23
#>
#> // tags: date_onset:date_onset, date_admission:date_admission, date_outcome:date_outcome
# Define dictionary
dat_dictionary <- tibble::tribble(
~options, ~values, ~grp, ~orders,
"1", "male", "sex", 1L,
"2", "female", "sex", 2L,
"M", "male", "sex", 3L,
"F", "female", "sex", 4L,
"m", "male", "sex", 5L,
"f", "female", "sex", 6L
)
# # Apply dictionary
# line_list <- line_list %>%
# cleanepi::clean_using_dictionary(
# dictionary = dat_dictionary
# )
# Very coverage of dictionary to solve the inconsistencies
line_list %>% count(sex)
#>
#> // linelist object
#> # A tibble: 15 × 2
#> sex n
#> <chr> <int>
#> 1 F 22
#> 2 Female 16
#> 3 M 15
#> 4 Male 19
#> 5 f 16
#> 6 female 14
#> 7 femvle 1
#> 8 femyle 1
#> 9 m 17
#> 10 malb 1
#> 11 mald 1
#> 12 male 9
#> 13 mmle 1
#> 14 myle 2
#> 15 <NA> 23
#>
#> // tags: date_onset:date_onset, date_admission:date_admission, date_outcome:date_outcome
# clean spelling mistakes using dictionary
line_list$case_type[agrep(pattern = "suspected", x = line_list$case_type)] <- "suspected"
line_list$case_type[agrep(pattern = "probable", x = line_list$case_type)] <- "probable"
line_list$case_type[agrep(pattern = "confirmed", x = line_list$case_type)] <- "confirmed"
line_list$outcome[agrep(pattern = "recovered", x = line_list$outcome)] <- "recovered"
line_list$outcome[agrep(pattern = "died", x = line_list$outcome)] <- "died"
# Validate clean line list ------------------------------------------------
# line list is now valid after cleaning
line_list_validated <- linelist::validate_linelist(line_list)
# Now, get data frame with tagged columns only
line_list_validated_tags <- linelist::tags_df(line_list_validated)
line_list_validated_tags
#> # A tibble: 158 × 3
#> date_onset date_admission date_outcome
#> <date> <date> <date>
#> 1 2023-01-01 2023-01-09 2023-01-13
#> 2 2023-01-01 NA NA
#> 3 NA NA NA
#> 4 2023-01-04 NA NA
#> 5 2023-01-05 2023-01-09 2023-01-23
#> 6 2023-01-06 2023-01-08 NA
#> 7 2023-01-13 NA NA
#> 8 2023-01-11 2023-01-24 2023-01-27
#> 9 NA NA NA
#> 10 2023-01-11 NA NA
#> # ℹ 148 more rows
# Aggregate and visualise data --------------------------------------------
# see visualising line list data vignette: https://epiverse-trace.github.io/simulist/articles/vis-linelist.html
# aggregate to daily incidence data
daily <- incidence2::incidence(
x = line_list_validated_tags,
date_index = "date_onset",
interval = "daily",
complete_dates = TRUE
)
# plot(daily)
# aggregate to epiweek incidence data
weekly <- incidence2::incidence(
x = line_list_validated_tags,
date_index = "date_onset",
interval = "epiweek",
complete_dates = TRUE
)
# plot(weekly)
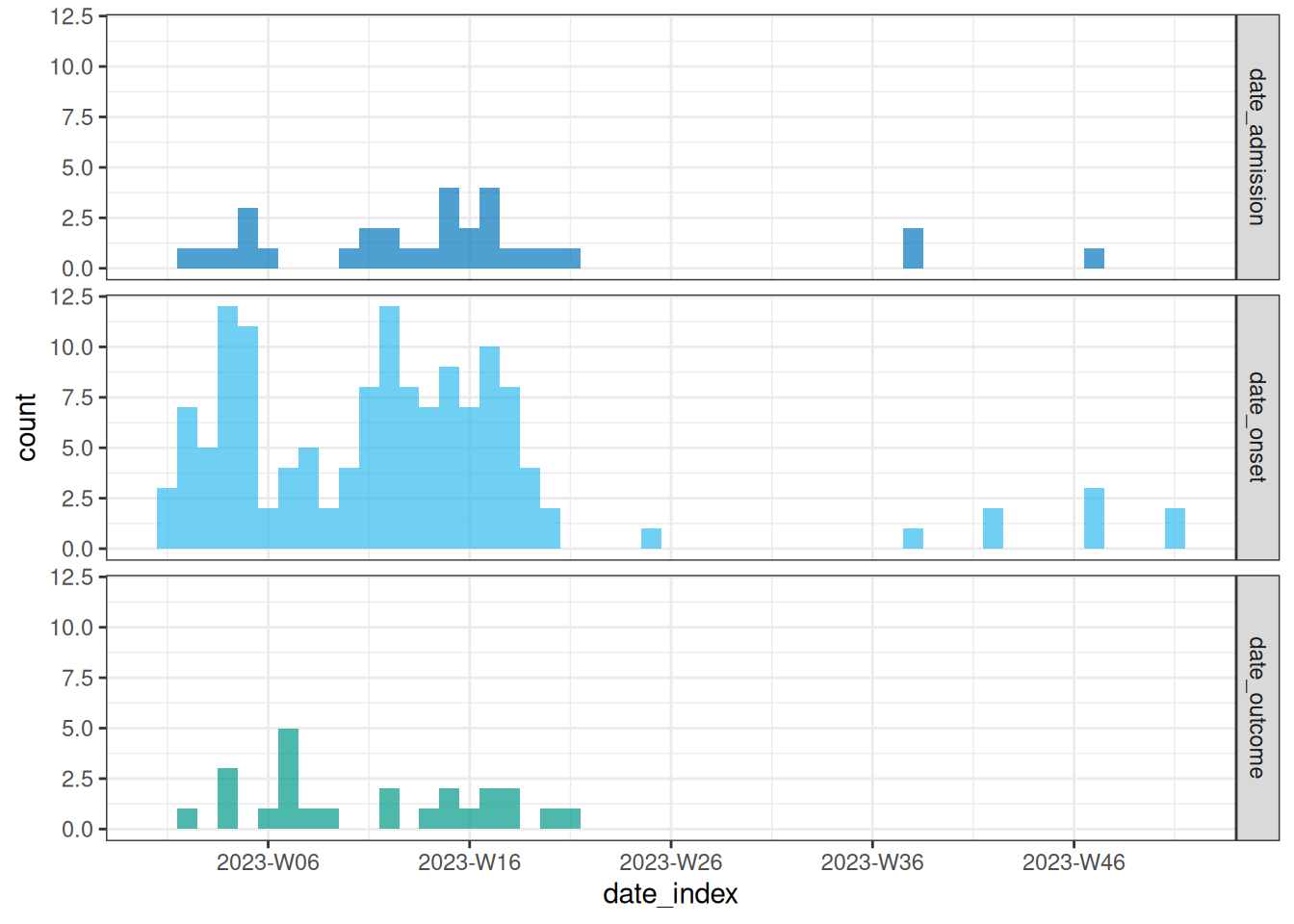
# aggregate and plot onset, hospital admission and death
weekly_chd <- line_list_validated_tags %>%
incidence2::incidence(
date_index = c("date_onset","date_admission","date_outcome"),
interval = "epiweek",
complete_dates = TRUE
)
plot(weekly_chd)