Appendix
Last updated on 2026-02-06 | Edit this page
Estimated time: 12 minutes
Overview
Questions
- Where can I add my functions?
- How do I need to document my functions?
- How can I read the documentation of my functions?
- How can I write a manuscript with my project outputs?
Objectives
- Write your functions documentation following the rcompendium template.
- Load your functions and update its documentation using devtools.
- Create a website for the project using usethis.
- Create a manuscript template with
{rrtools}.
What about my functions?
How do I write my functions documentation?
We must write our custom functions as Modular
functions and save them in the R/ folder. You can
write the documentation of your functions following a
standard documentation method.
The rcompendium template already contains a
fun-demo.R for this.
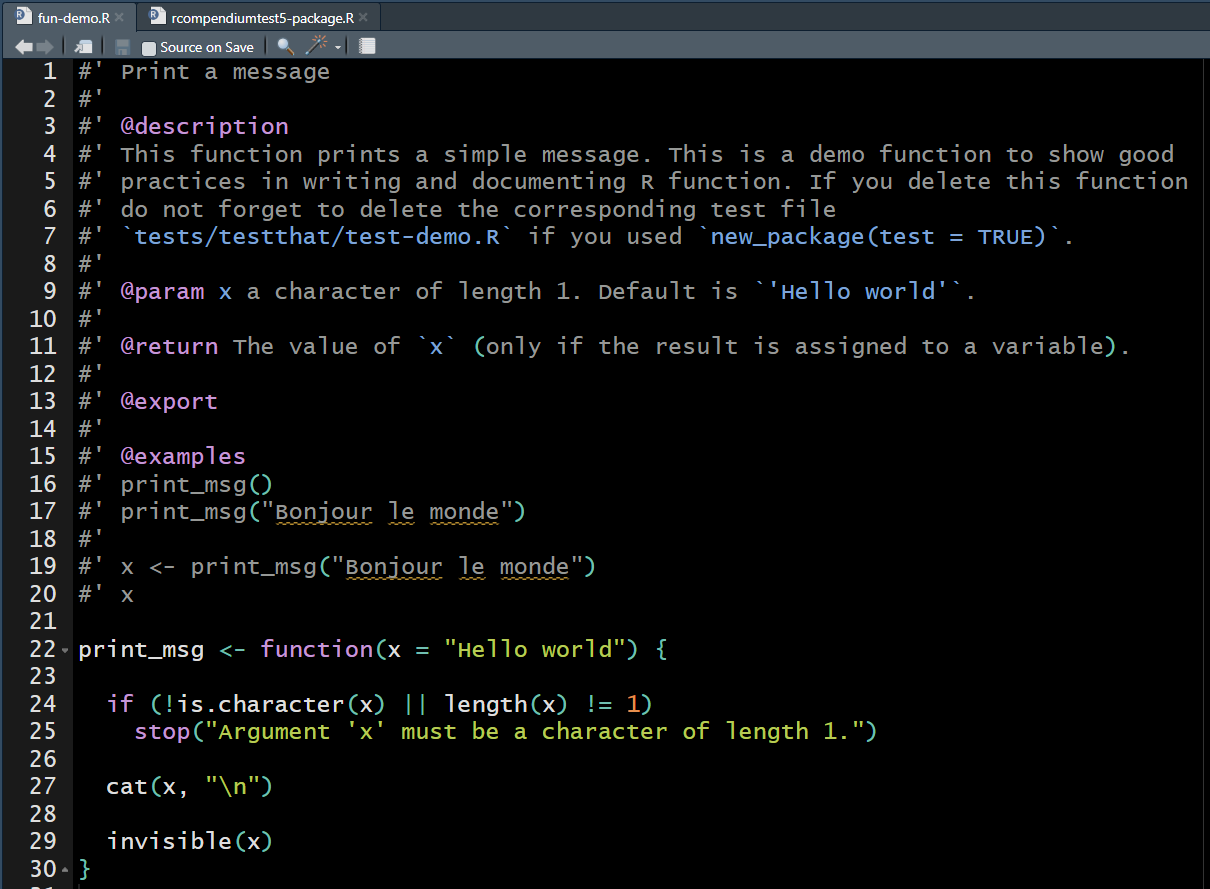
Callout
Remember that documented functions can facilitate further efforts to reuse them and create a specific R package!
How do I load my functions?
To load your project functions, as written in
line 20 of the make.R file, run:
R
devtools::load_all(here::here())
How do I read my functions documentation?
Is there an easier way to read the documentation of my modular functions?
Remember that after you write the documentation of new functions on
the R/ folder, you must update your function and project
documentation files, which are in different files and folders.
To do this run:
R
devtools::document()
This last step will update the following:
-
man/folder, which stores the project documentation, and - the
NAMESPACE, that registers the functions that your project exports for your data analysis to run.
Lastly, you can ask with ?function in the
Console and read the documentation for your functions, as
any other function from the packages you installed. Try to run this:
R
?print_msg
Create a project website
An alternative way to navigate all the files generated by the rcompendium template is with a website.
We can create a website using GitHub pages. To make this possible run:
R
usethis::use_pkgdown_github_pages()
This function implements the GitHub setup needed to automatically
publish your site to GitHub pages using the {pkgdown}
package.
This output is possible in two steps:
- First, it prepares to publish the pkgdown site from a new
gh-pagesbranch. - Then, it configures a GitHub Action to automatically build the site and deploy it via GitHub Pages.
Lastly, the pkgdown site’s URL is added to the
pkgdown configuration file, to the URL field of
DESCRIPTION, and to the GitHub repo.
Commit and Push your changes.
Callout
Remember that when using GitHub Actions, next to the
SHA/hash will be the status icon of the
actions.
- Yellow ball for “Job running”,
- Red cross for “Failed Run”, and
- Green check for “job done!”.
Please wait for it to get green and inspect the Reference tab on the navigation bar.
Now, let’s compare the fun-demo.R file, the
?print_msg output, and the website format:

Callout
A pkgdown website format can facilitate the navigation
through:
- Community files and
- Function documentation.
If required, you can add vignettes for an rcompendium using:
R
rcompendium::add_vignette(filename = "vignette-01")
Vignettes look more suitable for package documentation than a project. But knowing that we have that alternative with the rcompendium template is helpful.
How do I write a manuscript for my project?
You can use handy functions from another research compendium package
called {rrtools}.
To get a template of files required to fill a manuscript run:
R
rrtools::use_analysis(location = "inst", data_in_git = FALSE)
This function will create a folder inst/ with a new set
of folders for data and figures. You can avoid using them and only use
the .qmd as a template for your manuscript.
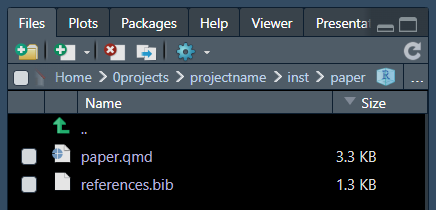
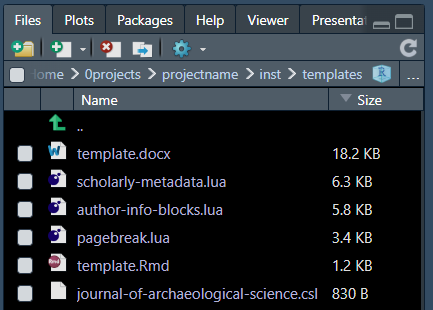
The .qmd files get formatted from several template files
like references using .bib and citation style using
.csl.
Using rrtools::use_analysis() with those arguments will
not modify your rcompendium configuration. Other
functions can change it.
This manuscript will not be visible on a website unless moved to the
vignette/ folder. We have yet to test that behaviour.
Reproducible research features
We also relate Reproducibility with the practice of describing and documenting the research process so that another researcher can re-run the software on the same data input to get the same data outputs.
Features related to this are:
Documentation strings in one or two lines using active verbs to describe how inputs turn into outputs (Irving et al. 2021). The documentation of functions, like the
fun-demo.Rtemplate file, follows this good practice.Manuscripts using literate programming with tools like Rmarkdown or Quarto. The template provided by
{rrtools}facilitates files to start with this practice.
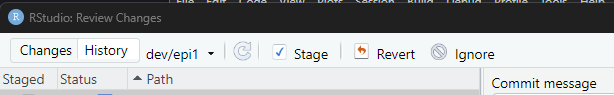
Callout
Remember that if you have all your changes as commits with git, you
can revert any modification with the button
Revert, located between the
Stage and Ignore buttons.
Key Points
- Write your functions documentation following the
R/fun-demo.Rtemplate. - Run your project functions with
devtools::load_all(). - Update your functions documentation with
devtools::document(). - Read your functions documentation with the
?functionnotation in the R console. - Create a website for the project with
usethis::use_pkgdown_github_pages(). - Use a manuscript template with
rrtools::use_analysis(location = "inst", data_in_git = FALSE). - Documentation strings and Manuscripts using literate programming are features related to Reproducible research.
