Introduction
Last updated on 2026-02-06 | Edit this page
Estimated time: 10 minutes
Overview
Questions
- Why to improve our code for analysis?
- What can we do to improve it?
- How can we start improving it?
Objectives
- Explain our vision of an improved epidemic analysis code.
- Share our strategy to incorporate good practices in scientific computing.
- Define our plan to incorporate practical and quick-to-learn solutions.
Why improve our code for epidemic analysis?
When we want to improve our analysis code’s reliability and reusability, we want to make it reproducible.
Reproducible research aims to ensure that anyone with access to data inputs and software can feasibly generate the data outputs, both to check or build on them. Reproducibility is improved when mixed with Open science and Sustainable software features.
Our vision for this workshop is to increase the awareness of good practices that will increase the reproducibility of data analysis workflow that already uses R and Git.
The figure above helps us to visualize and potentially evaluate the processes we are following. A process-centred approach helps us remove the focus on human error, be aware that processes can fail people with good intentions, and accept that we can enter a continuous improvement cycle.
“By defining the process, we can begin to borrow from the rich field of operations, which focuses primarily on (the) process. One paradigm that proves especially useful is the concept of human error. The seminal book The Field Guide to Understanding Human Error argues for a paradigm shift from the “Old World View” (that when an error occurs it is an individual actor’s fault) to the “New World View” (that when an error occurs, it is a symptom of a flawed system that failed that individual actor) (Dekker 2014). When an error in an analysis occurs, it is safe to assume (aside from nefarious actors) that the analyst did not want that error to occur. Given that she thought she was producing an analysis free from errors, you must look at the way she developed the analysis to understand where the error occurred, and create safeguards so that the error does not occur again.” (Parker, 2017a and Parker, 2017b)
Repetitive events (like outbreak response and research data analysis projects) give us the opportunity to:
- Focus on the process we have followed,
- Evaluate where bottlenecks occur, and then
- Adopt new practices to be better protected against errors in the next iteration.
Deming Cycle
This approach aims to follow a Deming cycle of Plan, Do, Check, and Act, as a foundation for continuous improvement.
Discussion
Exercise: Your experience analyzing outbreak data (the latest… or the most chaotic!)
Take 5 minutes.
Reflect on these questions:
- How do you organize your files and folders?
- Where do you describe what your project does or how to use it? Was it all in one accessible place?
- Could your project be reused by colleagues? Do you think it is?
Share one idea from your neighbour.
The questions above are self-assessment questions to let the participants hear and report back on their or others’ practices. This motivates a self-evaluation to include or replace previous practices, and potentially leave the session aware or motivated to proceed differently in the next iteration.
What can we do?
A fair strategy to follow is to gradually incorporate good practices in scientific computing (Wilson et al. 2017) that include:
- Data management,
- Software development,
- Collaboration,
- Project organization,
- Keep track of changes, and
- Manuscript writing.
Do I need to use them all from today?
No, we do not intend you start adopting all these workshop’s good practices and tools.
If you already use a programming language like R and Git for version control, you are already on the path!
We support the opinion of Jaime Quinn: “It can be challenging to absorb so many different good practices while still getting research done. However, I would argue that anything helps. While all good practices in open science are important, even just incorporating one example is good for the community and provides a solid personal foundation for gradually incorporating more good practices.”
How can we start?
Our plan for this workshop is to prioritize three tools, given their usefulness once mastered and the time to master them:
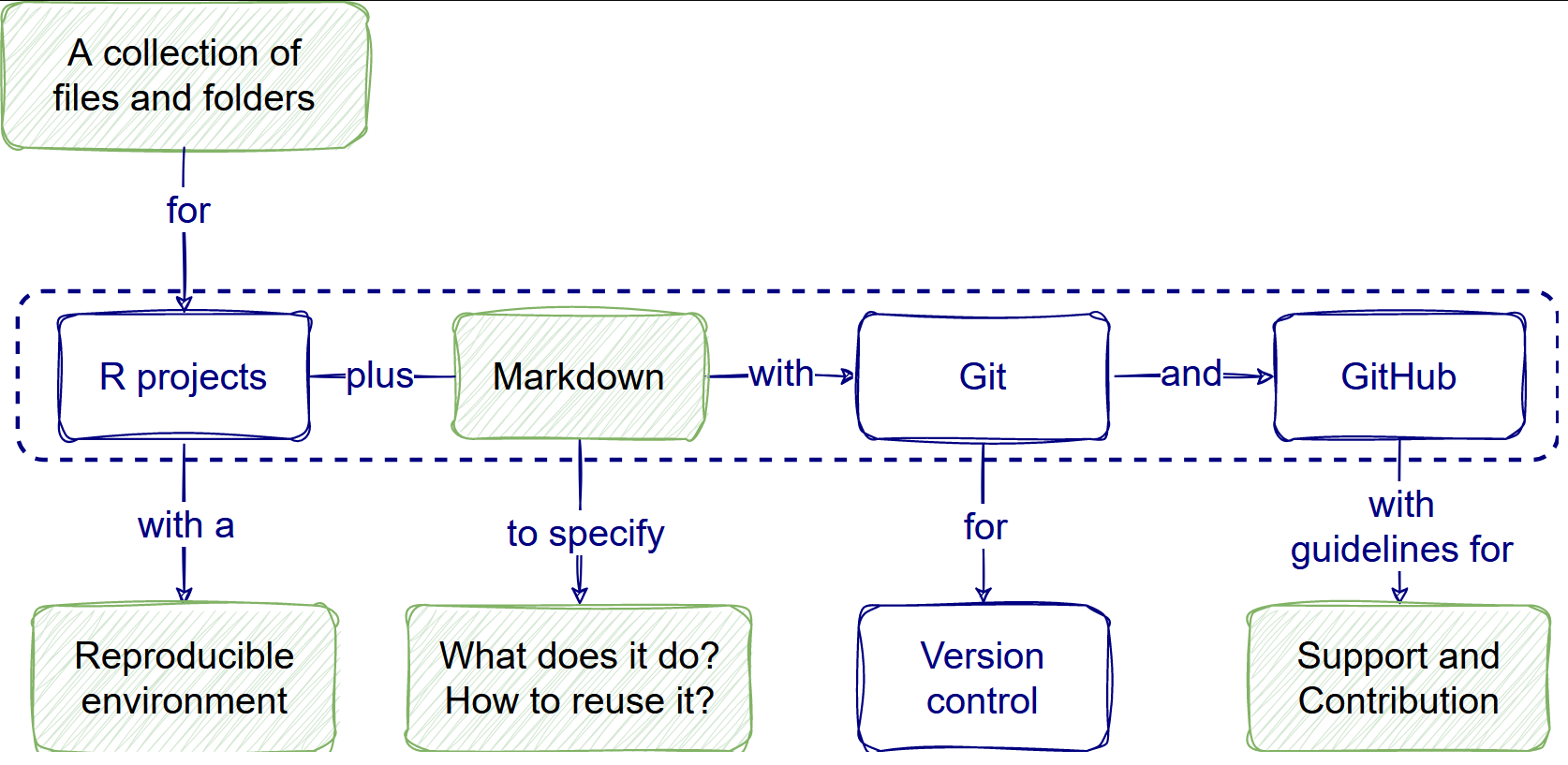
- Use research compendium templates.
- Make reproducible analysis.
- Write informative READMEs.
We’ll relate relevant features for Sustainable software, Reproducible research, and Open science for each tool.
Key Points
- Our vision is to increase the awareness of tools to improve the reproducibility of data analysis.
- Our strategy is to incorporate good practices in scientific computing gradually.
- We plan to share specific tools to create a research compendium, make a reproducible analysis, and write READMEs.
