Visualise results of the provided model
Usage
plot_seromodel(
seromodel,
serosurvey,
alpha = 0.05,
bin_serosurvey = FALSE,
bin_step = 5,
foi_df = NULL,
foi_max = NULL,
loo_estimate_digits = 1,
central_estimate_digits = 2,
seroreversion_digits = 2,
rhat_digits = 2,
size_text = 11,
plot_constant = FALSE,
x_axis = NA
)Arguments
- seromodel
stan_fit object obtained from sampling a model with fit_seromodel
- serosurvey
survey_yearYear in which the survey took place (only needed to plot time models)
age_minFloor value of the average between age_min and age_max
age_maxThe size of the sample
n_sampleNumber of samples for each age group
n_seropositiveNumber of positive samples for each age group
- alpha
1 - alpha indicates the credibility level to be used
- bin_serosurvey
If
TRUE,serodatais binned by means ofprepare_bin_serosurvey. Otherwise, age groups are kept as originally input.- bin_step
Integer specifying the age groups bin size to be used when
bin_serosurveyis set toTRUE.- foi_df
Dataframe with columns
year/ageYear/Age (depending on the model)
foiForce-of-infection values by year/age
- foi_max
Max FoI value for plotting
- loo_estimate_digits
Number of loo estimate digits
- central_estimate_digits
Number of central estimate digits
- seroreversion_digits
Number of seroreversion rate digits
- rhat_digits
Number of rhat estimate digits
- size_text
Size of text for plotting (
base_sizein ggplot2)- plot_constant
boolean specifying whether to plot single Force-of-Infection estimate and its corresponding rhat value instead of showing this information in the summary. Only relevant when
seromodel@model_name == "constant")- x_axis
either
"time"or"age". Specifies time axis values label for constant model additional plots. Only relevant when andseromodel@model_name == "constant"
Examples
# \donttest{
data(veev2012)
seromodel <- fit_seromodel(veev2012, iter = 100)
#>
#> SAMPLING FOR MODEL 'constant_no_seroreversion' NOW (CHAIN 1).
#> Chain 1: Rejecting initial value:
#> Chain 1: Log probability evaluates to log(0), i.e. negative infinity.
#> Chain 1: Stan can't start sampling from this initial value.
#> Chain 1:
#> Chain 1: Gradient evaluation took 1.7e-05 seconds
#> Chain 1: 1000 transitions using 10 leapfrog steps per transition would take 0.17 seconds.
#> Chain 1: Adjust your expectations accordingly!
#> Chain 1:
#> Chain 1:
#> Chain 1: WARNING: There aren't enough warmup iterations to fit the
#> Chain 1: three stages of adaptation as currently configured.
#> Chain 1: Reducing each adaptation stage to 15%/75%/10% of
#> Chain 1: the given number of warmup iterations:
#> Chain 1: init_buffer = 7
#> Chain 1: adapt_window = 38
#> Chain 1: term_buffer = 5
#> Chain 1:
#> Chain 1: Iteration: 1 / 100 [ 1%] (Warmup)
#> Chain 1: Iteration: 10 / 100 [ 10%] (Warmup)
#> Chain 1: Iteration: 20 / 100 [ 20%] (Warmup)
#> Chain 1: Iteration: 30 / 100 [ 30%] (Warmup)
#> Chain 1: Iteration: 40 / 100 [ 40%] (Warmup)
#> Chain 1: Iteration: 50 / 100 [ 50%] (Warmup)
#> Chain 1: Iteration: 51 / 100 [ 51%] (Sampling)
#> Chain 1: Iteration: 60 / 100 [ 60%] (Sampling)
#> Chain 1: Iteration: 70 / 100 [ 70%] (Sampling)
#> Chain 1: Iteration: 80 / 100 [ 80%] (Sampling)
#> Chain 1: Iteration: 90 / 100 [ 90%] (Sampling)
#> Chain 1: Iteration: 100 / 100 [100%] (Sampling)
#> Chain 1:
#> Chain 1: Elapsed Time: 0.003 seconds (Warm-up)
#> Chain 1: 0.002 seconds (Sampling)
#> Chain 1: 0.005 seconds (Total)
#> Chain 1:
#>
#> SAMPLING FOR MODEL 'constant_no_seroreversion' NOW (CHAIN 2).
#> Chain 2:
#> Chain 2: Gradient evaluation took 1.5e-05 seconds
#> Chain 2: 1000 transitions using 10 leapfrog steps per transition would take 0.15 seconds.
#> Chain 2: Adjust your expectations accordingly!
#> Chain 2:
#> Chain 2:
#> Chain 2: WARNING: There aren't enough warmup iterations to fit the
#> Chain 2: three stages of adaptation as currently configured.
#> Chain 2: Reducing each adaptation stage to 15%/75%/10% of
#> Chain 2: the given number of warmup iterations:
#> Chain 2: init_buffer = 7
#> Chain 2: adapt_window = 38
#> Chain 2: term_buffer = 5
#> Chain 2:
#> Chain 2: Iteration: 1 / 100 [ 1%] (Warmup)
#> Chain 2: Iteration: 10 / 100 [ 10%] (Warmup)
#> Chain 2: Iteration: 20 / 100 [ 20%] (Warmup)
#> Chain 2: Iteration: 30 / 100 [ 30%] (Warmup)
#> Chain 2: Iteration: 40 / 100 [ 40%] (Warmup)
#> Chain 2: Iteration: 50 / 100 [ 50%] (Warmup)
#> Chain 2: Iteration: 51 / 100 [ 51%] (Sampling)
#> Chain 2: Iteration: 60 / 100 [ 60%] (Sampling)
#> Chain 2: Iteration: 70 / 100 [ 70%] (Sampling)
#> Chain 2: Iteration: 80 / 100 [ 80%] (Sampling)
#> Chain 2: Iteration: 90 / 100 [ 90%] (Sampling)
#> Chain 2: Iteration: 100 / 100 [100%] (Sampling)
#> Chain 2:
#> Chain 2: Elapsed Time: 0.002 seconds (Warm-up)
#> Chain 2: 0.002 seconds (Sampling)
#> Chain 2: 0.004 seconds (Total)
#> Chain 2:
#>
#> SAMPLING FOR MODEL 'constant_no_seroreversion' NOW (CHAIN 3).
#> Chain 3:
#> Chain 3: Gradient evaluation took 1.3e-05 seconds
#> Chain 3: 1000 transitions using 10 leapfrog steps per transition would take 0.13 seconds.
#> Chain 3: Adjust your expectations accordingly!
#> Chain 3:
#> Chain 3:
#> Chain 3: WARNING: There aren't enough warmup iterations to fit the
#> Chain 3: three stages of adaptation as currently configured.
#> Chain 3: Reducing each adaptation stage to 15%/75%/10% of
#> Chain 3: the given number of warmup iterations:
#> Chain 3: init_buffer = 7
#> Chain 3: adapt_window = 38
#> Chain 3: term_buffer = 5
#> Chain 3:
#> Chain 3: Iteration: 1 / 100 [ 1%] (Warmup)
#> Chain 3: Iteration: 10 / 100 [ 10%] (Warmup)
#> Chain 3: Iteration: 20 / 100 [ 20%] (Warmup)
#> Chain 3: Iteration: 30 / 100 [ 30%] (Warmup)
#> Chain 3: Iteration: 40 / 100 [ 40%] (Warmup)
#> Chain 3: Iteration: 50 / 100 [ 50%] (Warmup)
#> Chain 3: Iteration: 51 / 100 [ 51%] (Sampling)
#> Chain 3: Iteration: 60 / 100 [ 60%] (Sampling)
#> Chain 3: Iteration: 70 / 100 [ 70%] (Sampling)
#> Chain 3: Iteration: 80 / 100 [ 80%] (Sampling)
#> Chain 3: Iteration: 90 / 100 [ 90%] (Sampling)
#> Chain 3: Iteration: 100 / 100 [100%] (Sampling)
#> Chain 3:
#> Chain 3: Elapsed Time: 0.004 seconds (Warm-up)
#> Chain 3: 0.002 seconds (Sampling)
#> Chain 3: 0.006 seconds (Total)
#> Chain 3:
#>
#> SAMPLING FOR MODEL 'constant_no_seroreversion' NOW (CHAIN 4).
#> Chain 4: Rejecting initial value:
#> Chain 4: Log probability evaluates to log(0), i.e. negative infinity.
#> Chain 4: Stan can't start sampling from this initial value.
#> Chain 4:
#> Chain 4: Gradient evaluation took 2.2e-05 seconds
#> Chain 4: 1000 transitions using 10 leapfrog steps per transition would take 0.22 seconds.
#> Chain 4: Adjust your expectations accordingly!
#> Chain 4:
#> Chain 4:
#> Chain 4: WARNING: There aren't enough warmup iterations to fit the
#> Chain 4: three stages of adaptation as currently configured.
#> Chain 4: Reducing each adaptation stage to 15%/75%/10% of
#> Chain 4: the given number of warmup iterations:
#> Chain 4: init_buffer = 7
#> Chain 4: adapt_window = 38
#> Chain 4: term_buffer = 5
#> Chain 4:
#> Chain 4: Iteration: 1 / 100 [ 1%] (Warmup)
#> Chain 4: Iteration: 10 / 100 [ 10%] (Warmup)
#> Chain 4: Iteration: 20 / 100 [ 20%] (Warmup)
#> Chain 4: Iteration: 30 / 100 [ 30%] (Warmup)
#> Chain 4: Iteration: 40 / 100 [ 40%] (Warmup)
#> Chain 4: Iteration: 50 / 100 [ 50%] (Warmup)
#> Chain 4: Iteration: 51 / 100 [ 51%] (Sampling)
#> Chain 4: Iteration: 60 / 100 [ 60%] (Sampling)
#> Chain 4: Iteration: 70 / 100 [ 70%] (Sampling)
#> Chain 4: Iteration: 80 / 100 [ 80%] (Sampling)
#> Chain 4: Iteration: 90 / 100 [ 90%] (Sampling)
#> Chain 4: Iteration: 100 / 100 [100%] (Sampling)
#> Chain 4:
#> Chain 4: Elapsed Time: 0.003 seconds (Warm-up)
#> Chain 4: 0.002 seconds (Sampling)
#> Chain 4: 0.005 seconds (Total)
#> Chain 4:
#> Warning: Bulk Effective Samples Size (ESS) is too low, indicating posterior means and medians may be unreliable.
#> Running the chains for more iterations may help. See
#> https://mc-stan.org/misc/warnings.html#bulk-ess
#> Warning: Tail Effective Samples Size (ESS) is too low, indicating posterior variances and tail quantiles may be unreliable.
#> Running the chains for more iterations may help. See
#> https://mc-stan.org/misc/warnings.html#tail-ess
plot_seromodel(seromodel, veev2012)
#> Warning: Some Pareto k diagnostic values are too high. See help('pareto-k-diagnostic') for details.
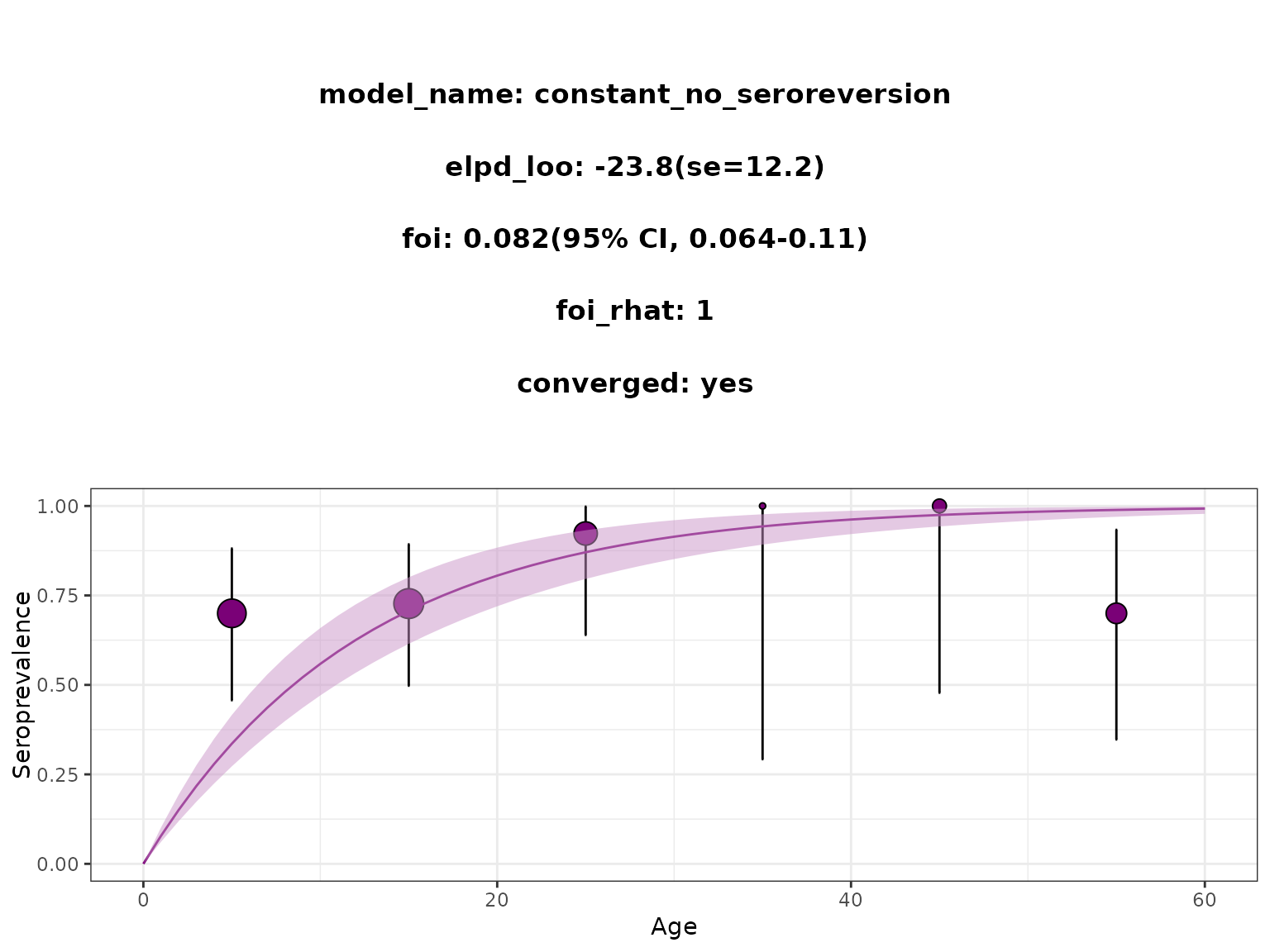 # }
# }
