Contact matrices
Last updated on 2026-02-05 | Edit this page
Overview
Questions
- What is a contact matrix?
- How are contact matrices estimated?
- How are contact matrices used in epidemiological analysis?
Objectives
- Use the R package
socialmixrto estimate a contact matrix - Understand the different types of analysis contact matrices can be used for
Introduction
Some groups of individuals have more contacts than others; the average schoolchild has many more daily contact than the average elderly person, for example. This heterogeneity of contact patterns between different groups can affect disease transmission, because certain groups are more likely to transmit to others within that group, as well as to other groups. The rate at which individuals within and between groups make contact with others can be summarised in a contact matrix. In this tutorial we are going to learn how contact matrices can be used in different analyses and how the socialmixr package can be used to estimate contact matrices.
R
library(socialmixr)
The contact matrix
The basic contact matrix represents the amount of contact or mixing within and between different subgroups of a population. The subgroups are often age categories but can also be:
- Geographic areas (e.g., different regions or countries)
- Risk groups (e.g., high/low risk occupations)
- Social settings (e.g., household, workplace, school)
For example, a hypothetical contact matrix representing the average number of contacts per day between children and adults could be:
\[ \begin{bmatrix} 2 & 2\\ 1 & 3 \end{bmatrix} \]
In this example, we would use this to represent that children meet, on average, 2 other children and 2 adult per day (first row), and adults meet, on average, 1 child and 3 other adults per day (second row). We can use this kind of information to account for the role heterogeneity in contact plays in infectious disease transmission.
A Note on Notation
For a contact matrix with rows \(i\) and columns \(j\):
- \(C[i,j]\) represents the average number of contacts that individuals in group \(i\) have with individuals in group \(j\)
- This average is calculated as the total number of contacts between groups \(i\) and \(j\), divided by the number of individuals in group \(i\)
Using socialmixr
Contact matrices are commonly estimated from studies that use diaries to record interactions. For example, the POLYMOD survey measured contact patterns in 8 European countries using data on the location and duration of contacts reported by the study participants (Mossong et al. 2008).
The R package socialmixr contains functions which can estimate contact matrices from POLYMOD and other surveys. We can load the POLYMOD survey data:
R
polymod <- socialmixr::polymod
Then we can obtain the contact matrix for the age categories we want
by specifying age_limits.
R
contact_data <- socialmixr::contact_matrix(
survey = polymod,
countries = "United Kingdom",
age_limits = c(0, 20, 40),
symmetric = TRUE
)
contact_data
OUTPUT
$matrix
contact.age.group
age.group [0,20) [20,40) 40+
[0,20) 7.883663 3.120220 3.063895
[20,40) 2.794154 4.854839 4.599893
40+ 1.565665 2.624868 5.005571
$demography
age.group population proportion year
<char> <num> <num> <int>
1: [0,20) 14799290 0.2454816 2005
2: [20,40) 16526302 0.2741283 2005
3: 40+ 28961159 0.4803901 2005
$participants
age.group participants proportion
<char> <int> <num>
1: [0,20) 404 0.3996044
2: [20,40) 248 0.2453017
3: 40+ 359 0.3550940Note: although the contact matrix
contact_data$matrix is not itself mathematically symmetric,
it satisfies the condition that the total number of contacts of one
group with another is the same as the reverse. In other words:
contact_data$matrix[j,i]*contact_data$demography$proportion[j] = contact_data$matrix[i,j]*contact_data$demography$proportion[i].
For the mathematical explanation see the
corresponding section in the socialmixr documentation.
Why would a contact matrix be non-symmetric?
One of the arguments we gave the function
contact_matrix() is symmetric=TRUE. This
ensures that the total number of contacts from one group to another is
equal to the total from the second group back to the first (see the
socialmixr vignette
for more detail).
However, when contact matrices are estimated from surveys or other sources, the reported number of contacts may differ by age group for several reasons:
- Recall bias: Different age groups may have different abilities to remember and report contacts accurately
- Reporting bias: Some groups may systematically over- or under-report their contacts
- Sampling uncertainty: Limited sample sizes can lead to statistical variations (Prem et al 2021)
If symmetric is set to TRUE, the
contact_matrix() function will internally use an average of
reported contacts to ensure the resulting total number of contacts are
symmetric.
The example above uses the POLYMOD survey. There are a number of
surveys available in socialmixr. To list the available
surveys, use socialmixr::list_surveys(). To download a
survey, we can use socialmixr::get_survey()
R
# Access the contact survey data from Zenodo
zambia_sa_survey <- socialmixr::get_survey(
"https://doi.org/10.5281/zenodo.3874675"
)
You can explore all the available surveys from the Zenodo repository at https://zenodo.org/communities/social_contact_data/. If you are interested in accessing to a specific URL within R, you can try:
R
library(socialmixr)
library(tidyverse)
# Get URL for Zambia contact survey data from {socialmixr}
socialmixr::list_surveys() %>%
dplyr::filter(stringr::str_detect(title, "Zambia")) %>%
dplyr::pull(url)
Zambia contact matrix
The R package {socialmixr} contains functions which can estimate contact matrices from POLYMOD and other surveys. Outputs include demographic information like population size and number of participants in the study. Using {socialmixr}:
Get access to the survey from Zambia.
-
Generate a symmetric contact matrix for Zambia using the following age bins:
- [0,20)
- 20+
Get access to the vector of
populationsize per age bin from thedemographydataset inside the contact matrix output.
The survey object zambia_sa_survey contains data from
two countries. If you need to estimate the social contact matrix from
data of the specific country of Zambia, identify what argument in
socialmixr::contact_matrix() you need for this.
R
# Inspect the countries within the survey object
levels(zambia_sa_survey$participants$country)
OUTPUT
[1] "South Africa" "Zambia" Similar to the code above, to access vector values within a
dataframe, you can use the dollar-sign operator: $
Synthetic contact matrices
Contact matrices can be estimated from data obtained from diary (such as POLYMOD), survey or contact data, or synthetic ones can be used. Prem et al. 2021 used the POLYMOD data within a Bayesian hierarchical model to project contact matrices for 177 other countries.
Analyses with contact matrices
Contact matrices can be used in a wide range of epidemiological analyses, they can be used:
- to calculate the basic reproduction number while accounting for different rates of contacts between age groups (Funk et al. 2019),
- to calculate final size of an epidemic, as in the R package finalsize,
- to assess the impact of interventions finding the relative change between pre and post intervention contact matrices to calculate the relative difference in \(R_0\) (Jarvis et al. 2020),
- and in mathematical models of transmission within a population, to account for group specific contact patterns.
However, all of these applications require us to perform some additional calculations using the contact matrix. Specifically, there are two main calculations we often need to do:
- Convert contact matrix into expected number of secondary cases
If contacts vary between groups, then the average number of secondary
cases won’t be equal simply to the average number of contacts multiplied
by the probability of transmission-per-contact. This is because the
average amount of transmission in each generation of infection isn’t
just a matter of whom a group came into contact with; it’s about whom
their contacts subsequently come into contact with. The
function r_eff in the package finalsize can
perform this conversion, taking a contact matrix, demography and
proportion susceptible and converting it into an estimate of the average
number of secondary cases generated by a typical infectious individual
(i.e. the effective reproduction number).
- Convert contact matrix into contact rates
Whereas a contact matrix gives the average number of contacts that one groups makes with another, epidemic dynamics in different groups depend on the rate at which one group infects another. We therefore need to scale the rate of interaction between different groups (i.e. the number of contacts per unit time) to get the rate of transmission. However, we need to be careful that we are defining transmission to and from each group correctly in any model. Specifically, the entry \((i,j)\) in a mathematical model contact matrix represents contacts of group \(i\) with group \(j\). But if we want to know the rate at which a group \(i\) are getting infected, then we want to multiply the number of contacts of susceptibles in group \(i\) (\(S_i\)) with group \(j\) (\(C[i,j]\)) with the proportion of those contacts that are infectious (\(I_j/N_j\)), and the transmission risk per contact (\(\beta\)).
In mathematical models
Consider the SIR model where individuals are categorized as either susceptible \(S\), infected but not yet infectious \(E\), infectious \(I\) or recovered \(R\). The schematic below shows the processes which describe the flow of individuals between the disease states \(S\), \(I\) and \(R\) and the key parameters for each process.
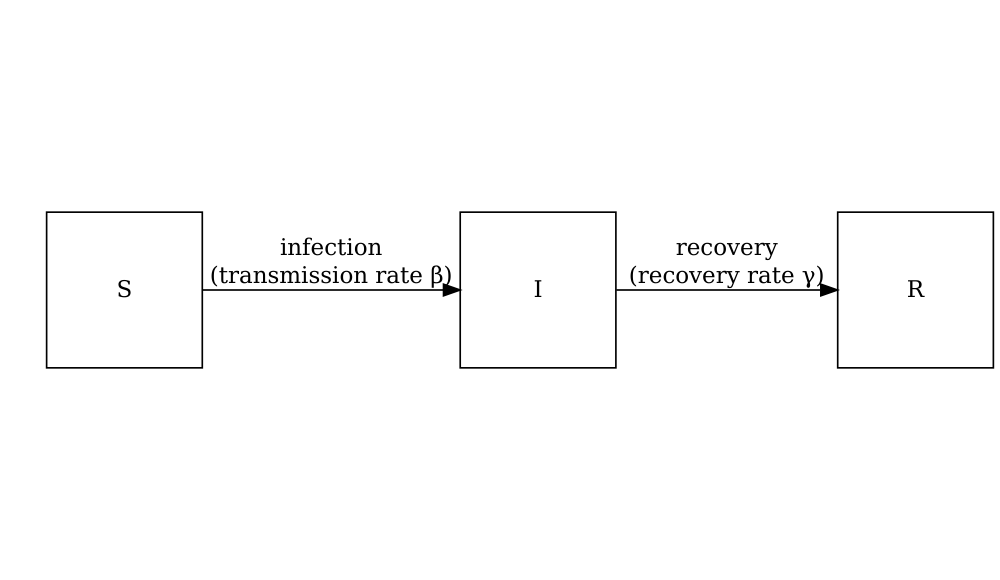
The differential equations below describe how individuals move from one state to another (Bjørnstad et al. 2020).
\[ \begin{aligned} \frac{dS}{dt} & = - \beta S I /N \\ \frac{dI}{dt} &= \beta S I /N - \gamma I \\ \frac{dR}{dt} &=\gamma I \\ \end{aligned} \]
To add age structure to our model, we need to add additional equations for the infection states \(S\), \(I\) and \(R\) for each age group \(i\). If we want to assume that there is heterogeneity in contacts between age groups then we must adapt the transmission term \(\beta SI\) to include the contact matrix \(C\) as follows :
\[ \beta S_i \sum_j C_{i,j} I_j/N_j. \]
Susceptible individuals in age group \(i\) become infected dependent on their rate of contact with individuals in each age group. For each disease state (\(S\), \(E\), \(I\) and \(R\)) and age group (\(i\)), we have a differential equation describing the rate of change with respect to time.
\[ \begin{aligned} \frac{dS_i}{dt} & = - \beta S_i \sum_j C_{i,j} I_j/N_j \\ \frac{dI_i}{dt} &= \beta S_i\sum_j C_{i,j} I_j/N_j - \gamma I_i \\ \frac{dR_i}{dt} &=\gamma I_i \\ \end{aligned} \]
Normalising the contact matrix to ensure the correct value of \(R_0\)
When simulating an epidemic, we often want to ensure that the average number of secondary cases generated by a typical infectious individual (i.e. \(R_0\)) is consistent with known values for the pathogen we’re analysing. In the above model, we scale the contact matrix by the \(\beta\) to convert the raw interaction data into a transmission rate. But how do we define the value of \(\beta\) to ensure a certain value of \(R_0\)?
Rather than just using the raw number of contacts, we can instead normalise the contact matrix to make it easier to work in terms of \(R_0\). In particular, we normalise the matrix by scaling it so that if we were to calculate the average number of secondary cases based on this normalised matrix, the result would be 1 (in mathematical terms, we are scaling the matrix so the largest eigenvalue is 1). This transformation scales the entries but preserves their relative values.
In the case of the above model, we want to define \(\beta C_{i,j}\) so that the model has a
specified valued of \(R_0\). If the
entry of the contact matrix \(C[i,j]\)
represents the contacts of population \(i\) with \(j\), it is equivalent to
contact_data$matrix[i,j], and the maximum eigenvalue of
this matrix represents the typical magnitude of contacts, not typical
magnitude of transmission. We must therefore normalise the matrix \(C\) so the maximum eigenvalue is one; we
call this matrix \(C_{normalised}\).
Because the rate of recovery is \(\gamma\), individuals will be infectious on
average for \(1/\gamma\) days. So \(\beta\) as a model input is calculated from
\(R_0\), the scaling factor and the
value of \(\gamma\)
(i.e. mathematically we use the fact that the dominant eigenvalue of the
matrix \(R_0 \times C_{normalised}\) is
equal to \(\beta / \gamma\)).
R
contact_matrix <- t(contact_data$matrix)
scaling_factor <- 1 / max(eigen(contact_matrix)$values)
normalised_matrix <- contact_matrix * scaling_factor
As a result, if we multiply the scaled matrix by \(R_0\), then converting to the number of expected secondary cases would give us \(R_0\), as required.
R
infectious_period <- 7.0
basic_reproduction <- 1.46
transmission_rate <- basic_reproduction * scaling_factor / infectious_period
# check the dominant eigenvalue of R0 x C_normalised is R0
max(eigen(basic_reproduction * normalised_matrix)$values)
OUTPUT
[1] 1.46Check the dimension of \(\beta\)
In the SIR model without age structure the rate of contact is part of the transmission rate \(\beta\), where as in the age-structured model we have separated out the rate of contact, hence the transmission rate \(\beta\) in the age structured model will have a different value.
We can use contact matrices from socialmixr with
mathematical models in the R package {epidemics}. See the
tutorial Simulating
transmission for examples and an introduction to
epidemics.
Contact groups
In the example above the dimension of the contact matrix will be the same as the number of age groups i.e. if there are 3 age groups then the contact matrix will have 3 rows and 3 columns. Contact matrices can be used for other groups as long as the dimension of the matrix matches the number of groups.
For example, we might have a meta population model with two geographic areas. Then our contact matrix would be a 2 x 2 matrix with entries representing the contact between and within the geographic areas.
Summary
In this tutorial, we have learnt the definition of the contact
matrix, how they are estimated and how to access social contact data
from socialmixr. In the next tutorial, we will learn how to
use the R package {epidemics} to generate disease
trajectories from mathematical models with contact matrices from
socialmixr.
Key Points
- Contact matrices quantify the mixing patterns between different population groups
-
socialmixrprovides tools to estimate contact matrices from survey data - Contact matrices can be used in various epidemiological analyses, from calculating \(R_0\) to modeling interventions
- Proper normalization is crucial when using contact matrices in transmission models
