
Using {epireview} with {epiparameter}
Source:vignettes/articles/data_from_epireview.Rmd
data_from_epireview.Rmd
library(epiparameter)
library(epireview)
#> Loading required package: epitrix
#> Loading required package: ggplot2
#> Loading required package: ggforceThe article describes how to use the {epireview} R package with
{epiparameter}. {epireview} provides epidemiological parameters for a
range of pathogens extracted from the literature by the Pathogen
Epidemiology Review Group (PERG) in systematic reviews. The
{epiparameter} R package can use these epidemiological parameters and
can convert them into <epiparameter> objects.
This feature of interoperability of {epireview} and {epiparameter} is
still under development. Currently only delay distributions (termed
Human delay in the “parameter_type” column in {epireview}’s
database) are compatible with this feature. Attempting to convert other
epidemiological parameters from {epireview} to {epiparameter}
<epiparameter> objects will likely error with an
uninformative error message.
Converting from {epireview} entries into an
<epiparameter> object
The {epireview} package nicely provides epidemiological parameter
data from systematically reviewing the literature, and {epiparameter}
provides custom data structures for working with epidemiological data in
R. Therefore, reading in data from the {epireview} R package and
converting it to an <epiparameter> object will
provide the greatest utility when applied in outbreak analytics.
Here we start with a simple example of reading in the Marburg data
from {epireview} and converting to an <epiparameter>
object using as_epiparameter() function from the
{epiparameter} package.
marburg_data <- load_epidata("marburg")
#> Warning: There is 1 article with missing first author surname.
#> Warning: There is 1 article with missing first author surname and first author first
#> name.
#> Warning: There is 1 article with missing year of publication.
#> Warning: Unknown or uninitialised column: `other_delay_start`.
#> Warning: Unknown or uninitialised column: `other_delay_end`.
#> Note: the params dataframe does not have a covidence_id column
#> Note: the models dataframe does not have a covidence_id column
#> Note: the outbreaks dataframe does not have a covidence_id column
#> ✔ Data loaded for marburgThis loads a list with four tables, specifically
tibbles, that contain the bibliographic information
($articles), epidemiological parameters
($params), epidemiological models ($models),
and outbreak information ($outbreaks).
names(marburg_data)
#> [1] "articles" "params" "models" "outbreaks"We will start by just using the epidemiological parameter table to
convert information into an <epiparameter>.
marburg_params <- marburg_data$paramsOut of these parameters, subset the data to only keep those rows that contain incubation periods for Marburg.
marburg_incubation_period <- marburg_params[
marburg_params$parameter_type_short == "incubation_period",
]
marburg_incubation_period
#> # A tibble: 2 × 61
#> parameter_data_id article_id parameter_type parameter_value parameter_unit
#> <chr> <int> <chr> <dbl> <chr>
#> 1 c2a35e68034b72580654… 6 Human delay -… NA Days
#> 2 0106582cf5ed3c52d5e9… 20 Human delay -… NA Days
#> # ℹ 56 more variables: parameter_lower_bound <dbl>,
#> # parameter_upper_bound <dbl>, parameter_value_type <chr>,
#> # parameter_uncertainty_single_value <dbl>,
#> # parameter_uncertainty_singe_type <chr>,
#> # parameter_uncertainty_lower_value <dbl>,
#> # parameter_uncertainty_upper_value <dbl>, parameter_uncertainty_type <chr>,
#> # cfr_ifr_numerator <int>, cfr_ifr_denominator <int>, …We will select the first entry to use as the first example:
marburg_incub <- marburg_incubation_period[1, ]
marburg_incub
#> # A tibble: 1 × 61
#> parameter_data_id article_id parameter_type parameter_value parameter_unit
#> <chr> <int> <chr> <dbl> <chr>
#> 1 c2a35e68034b72580654… 6 Human delay -… NA Days
#> # ℹ 56 more variables: parameter_lower_bound <dbl>,
#> # parameter_upper_bound <dbl>, parameter_value_type <chr>,
#> # parameter_uncertainty_single_value <dbl>,
#> # parameter_uncertainty_singe_type <chr>,
#> # parameter_uncertainty_lower_value <dbl>,
#> # parameter_uncertainty_upper_value <dbl>, parameter_uncertainty_type <chr>,
#> # cfr_ifr_numerator <int>, cfr_ifr_denominator <int>, …Then we can simply pass our epidemiological parameter set to
as_epiparameter() to do the conversion.
marburg_incub_epiparameter <- as_epiparameter(marburg_incub)
#> Using Gear (1975). "<title not available>." _<journal not available>_.
#> To retrieve the citation use the 'get_citation' function
#> Warning: Cannot create full citation for epidemiological parameters without bibliographic information
#> see ?as_epiparameter for help.
#> No adequate summary statistics available to calculate the parameters of the NA distribution
#> Unparameterised <epiparameter> object
marburg_incub_epiparameter
#> Disease: Marburg Virus Disease
#> Pathogen: Marburg virus
#> Epi Parameter: human delay incubation period
#> Study: Gear (1975). "<title not available>." _<journal not available>_.
#> Distribution: NA
#> Mean: NA [NA% CI: NA, NA] (Days)
#> Median: NA [NA% CI: NA, NA] (Days)
#> Range: [c(7, 8)] (Days)The resulting <epiparameter> does not contain a
parameterised probability distribution, instead it contains a range for
the incubation period (in $summary_stats), and the
$metadata shows that this is a single case from South
Africa.
marburg_incub_epiparameter$summary_stats
#> $mean
#> [1] NA
#>
#> $mean_ci_limits
#> [1] NA NA
#>
#> $mean_ci
#> [1] NA
#>
#> $sd
#> [1] NA
#>
#> $sd_ci_limits
#> [1] NA NA
#>
#> $sd_ci
#> [1] NA
#>
#> $median
#> [1] NA
#>
#> $median_ci_limits
#> [1] NA NA
#>
#> $median_ci
#> [1] NA
#>
#> $dispersion
#> [1] NA
#>
#> $dispersion_ci_limits
#> [1] NA NA
#>
#> $dispersion_ci
#> [1] NA
#>
#> $quantiles
#> [1] NA
#>
#> $range
#> [1] 7 8
marburg_incub_epiparameter$metadata
#> $units
#> [1] "Days"
#>
#> $sample_size
#> [1] 1
#>
#> $region
#> [1] "Johannesburg, South Africa"
#>
#> $transmission_mode
#> [1] NA
#>
#> $vector
#> [1] NA
#>
#> $extrinsic
#> [1] FALSE
#>
#> $inference_method
#> [1] NACreating an <epiparameter> with full
citation
The last example showed how to convert the epidemiological parameter information, however, you may have noticed that the citation that was created did not contain the information for a full citation.
marburg_incub_epiparameter$citation
#> Gear (1975). "<title not available>." _<journal not available>_.In order to provide a complete citation to the
<epiparameter> object, which is highly recommended so
that you know the provenance of the parameters and can correctly
attribute the original authors, we will need to also provide the
bibliographic information from {epireview} as well as the
epidemiological parameters.
The article data needs to be loaded from {epireview} using
epireview::load_epidata_raw() rather than
epireview::load_data() because load_data()
subsets the bibliographic information to only provide:
"id", "first_author_surname",
"year_publication", and "article_label"
columns.
marburg_articles <- load_epidata_raw(
pathogen = "marburg",
table = "article"
)
marburg_articles
#> # A tibble: 58 × 25
#> article_id pathogen covidence_id first_author_first_n…¹ article_title doi
#> <dbl> <chr> <int> <chr> <chr> <chr>
#> 1 1 Marburg v… 2059 G A Haemorrhagic… NA
#> 2 2 Marburg v… 2042 Christian Antibodies t… NA
#> 3 3 Marburg v… 1649 Y The origin a… 10.1…
#> 4 4 Marburg v… 1692 D.H. Marburg-Viru… NA
#> 5 5 Marburg v… 2597 E. D. Filovirus ac… NA
#> 6 6 Marburg v… 3795 JS Outbreak of … 10.1…
#> 7 7 Marburg v… 2596 E.D. Haemorrhagic… NA
#> 8 8 Marburg v… 1615 O Viral hemorr… 10.4…
#> 9 9 Marburg v… 1693 Smiley Suspected Ex… 10.1…
#> 10 10 Marburg v… 1692 D Marburg-viru… NA
#> # ℹ 48 more rows
#> # ℹ abbreviated name: ¹first_author_first_name
#> # ℹ 19 more variables: journal <chr>, year_publication <int>, volume <int>,
#> # issue <int>, page_first <int>, page_last <int>, paper_copy_only <lgl>,
#> # notes <chr>, first_author_surname <chr>, double_extracted <dbl>,
#> # qa_m1 <chr>, qa_m2 <chr>, qa_a3 <chr>, qa_a4 <chr>, qa_d5 <chr>,
#> # qa_d6 <chr>, qa_d7 <chr>, score <dbl>, id <chr>We need to match the entry in the epidemiological parameter table with the citation information in the article table to ensure we are using the correct citation for the parameter set. Thankfully, this can easily be achieved as {epireview} provides unique IDs to each table to link entries.
article_row <- match(marburg_incub$id, marburg_articles$id)
article_row
#> [1] 6
marburg_incub_article <- marburg_articles[article_row, ]
marburg_incub_article
#> # A tibble: 1 × 25
#> article_id pathogen covidence_id first_author_first_n…¹ article_title doi
#> <dbl> <chr> <int> <chr> <chr> <chr>
#> 1 6 Marburg vi… 3795 JS Outbreak of … 10.1…
#> # ℹ abbreviated name: ¹first_author_first_name
#> # ℹ 19 more variables: journal <chr>, year_publication <int>, volume <int>,
#> # issue <int>, page_first <int>, page_last <int>, paper_copy_only <lgl>,
#> # notes <chr>, first_author_surname <chr>, double_extracted <dbl>,
#> # qa_m1 <chr>, qa_m2 <chr>, qa_a3 <chr>, qa_a4 <chr>, qa_d5 <chr>,
#> # qa_d6 <chr>, qa_d7 <chr>, score <dbl>, id <chr>Now we can repeat the example of converting to
<epiparameter> as shown above, but this time pass the
bibliographic information as well as the epidemiological parameter
information to create a full citation. The bibliographic information
needs to be passed with the article argument.
marburg_incub_epiparameter <- as_epiparameter(
marburg_incub,
article = marburg_incub_article
)
#> Using Gear (1975). "Outbreak of Marburg virus disease in Johannesburg." _The
#> British Medical Journal_. doi:10.1136/bmj.4.5995.489
#> <https://doi.org/10.1136/bmj.4.5995.489>.
#> To retrieve the citation use the 'get_citation' function
#> No adequate summary statistics available to calculate the parameters of the NA distribution
#> Unparameterised <epiparameter> object
marburg_incub_epiparameter
#> Disease: Marburg Virus Disease
#> Pathogen: Marburg virus
#> Epi Parameter: human delay incubation period
#> Study: Gear (1975). "Outbreak of Marburg virus disease in Johannesburg." _The
#> British Medical Journal_. doi:10.1136/bmj.4.5995.489
#> <https://doi.org/10.1136/bmj.4.5995.489>.
#> Distribution: NA
#> Mean: NA [NA% CI: NA, NA] (Days)
#> Median: NA [NA% CI: NA, NA] (Days)
#> Range: [c(7, 8)] (Days)
marburg_incub_epiparameter$citation
#> Gear (1975). "Outbreak of Marburg virus disease in Johannesburg." _The
#> British Medical Journal_. doi:10.1136/bmj.4.5995.489
#> <https://doi.org/10.1136/bmj.4.5995.489>.The as_epiparameter() function is an S3 generic. If you
are not familiar with S3 object-oriented programming in R, then this
detail is not important, however, it does mean that the
article argument is not explicitly in the function
definition of as_epiparameter() (i.e. it will not show up
on autocomplete when typing out the function and will not be shown if
you read the function help page ?as_epiparameter()).
Instead, the argument is specified as part of the ...
argument. This is because the article argument is only
required when converting data from {epireview} into
<epiparameter>, and other data that can be converted
to <epiparameter> objects do not require this
argument.
Multi-row {epireview} entries
In general, all required values for a parameter are represented as a single entry in epireview. In some of cases, e.g. for Marburg Virus Disease and Ebola Virus Disease (as the first pathogens the PERG team extracted), values captured for a parameter are on multiple rows. We are trying to avoid this as linking these entries is very challenging, but there are still some cases in which there are linked parameters on different rows. We provide further information in the limitations section below.
The way the {epireview} data is stored means that some
epidemiological parameter entries require multiple rows. This can be,
for example, because they contain two summary statistics (e.g. mean and
standard deviation) that are kept on separate rows. In order to create
<epiparameter> objects that contains the full
information for each entry multiple rows of the epidemiological
parameters table from {epireview} can be given to
as_epiparameter() to create a single
<epiparameter> object.
We can search which entries in the data have multiple rows by checking if there are duplicated parameter types and IDs. Remember that it is only possible to convert delay distributions into epiparameter objects (i.e. known as Human delay parameter types in {epireview}).
multi_row_entries <- duplicated(marburg_params$parameter_type) &
duplicated(marburg_params$id)
multi_row_ids <- marburg_params$id[multi_row_entries]
multi_row_marburg_params <-
marburg_params[marburg_params$id %in% multi_row_ids, ]
multi_row_marburg_params
#> # A tibble: 42 × 61
#> parameter_data_id article_id parameter_type parameter_value parameter_unit
#> <chr> <int> <chr> <dbl> <chr>
#> 1 0106582cf5ed3c52d5e… 20 Human delay -… NA Days
#> 2 ce78f707a585d8bb23a… 22 Seroprevalenc… 0 Percentage (%)
#> 3 ca720828fd6ccb18844… 22 Seroprevalenc… 0 NA
#> 4 61fbb9dfc021abf5bd1… 22 Seroprevalenc… 0 Percentage (%)
#> 5 29c8ca74306713a990c… 20 Severity - ca… NA NA
#> 6 056a8d6b5f9aee3622d… 27 Human delay -… 9 Days
#> 7 ce3976e2e15df3f6fb9… 27 Human delay -… 5.4 Days
#> 8 3bf73665fa67a6ba7f7… 27 Human delay -… 7 Days
#> 9 ba019f18acac9c5b0b7… 27 Human delay -… 9.3 Days
#> 10 71798b4154011dcd008… 27 Human delay -… 9 Days
#> # ℹ 32 more rows
#> # ℹ 56 more variables: parameter_lower_bound <dbl>,
#> # parameter_upper_bound <dbl>, parameter_value_type <chr>,
#> # parameter_uncertainty_single_value <dbl>,
#> # parameter_uncertainty_singe_type <chr>,
#> # parameter_uncertainty_lower_value <dbl>,
#> # parameter_uncertainty_upper_value <dbl>, …In this case there are two studies for Marburg with more than one entry (row) in the {epireview} database. Out of these studies we select the mean and standard deviation.
multi_row_marburg_params$parameter_value_type
#> [1] NA NA NA
#> [4] NA NA "Mean"
#> [7] "Standard Deviation" "Median" "Mean"
#> [10] "Median" "Mean" "Mean"
#> [13] "Mean" "Mean" "Mean"
#> [16] NA NA NA
#> [19] NA "Mean" "Mean"
#> [22] "Mean" "Mean" NA
#> [25] NA "Median" NA
#> [28] NA NA NA
#> [31] "Other" NA NA
#> [34] NA "Other" "Other"
#> [37] "Other" "Other" "Other"
#> [40] "Other" "Mean" "Other"In this case, we know that the mean and standard deviation from the
chosen rows correspond to the same estimation process by having read the
corresponding article. However, there is currently no identifiers on the
{epireview} params database for Marburg, Ebola or Lassa to
directly identify which of the two rows with mean values correspond to
the standard deviation. The {epireview} team are currently working on
rectifying this issue.
Therefore, we encourage readers to manually verify their data subsets, to ensure that the entries that have been selected are indeed multiple rows for the same reported epidemiological parameter.
- For future {epireview} pathogens (excluding SARS) mean and standard
deviation estimates that match will form one row in the
$paramsdatabase. Current software development at {epireview} is working on ensuring compatibility between these formats.
marburg_gt <- multi_row_marburg_params[
multi_row_marburg_params$parameter_data_id %in%
c("056a8d6b5f9aee3622d3bd8b715d4296", "ce3976e2e15df3f6fb92f6deb2db2a29"),
]
marburg_gt
#> # A tibble: 2 × 61
#> parameter_data_id article_id parameter_type parameter_value parameter_unit
#> <chr> <int> <chr> <dbl> <chr>
#> 1 056a8d6b5f9aee3622d3… 27 Human delay -… 9 Days
#> 2 ce3976e2e15df3f6fb92… 27 Human delay -… 5.4 Days
#> # ℹ 56 more variables: parameter_lower_bound <dbl>,
#> # parameter_upper_bound <dbl>, parameter_value_type <chr>,
#> # parameter_uncertainty_single_value <dbl>,
#> # parameter_uncertainty_singe_type <chr>,
#> # parameter_uncertainty_lower_value <dbl>,
#> # parameter_uncertainty_upper_value <dbl>, parameter_uncertainty_type <chr>,
#> # cfr_ifr_numerator <int>, cfr_ifr_denominator <int>, …We can now convert this to an <epiparameter>.
marburg_gt_epiparameter <- as_epiparameter(marburg_gt)
#> Using Ajelli (2012). "<title not available>." _<journal not available>_.
#> To retrieve the citation use the 'get_citation' function
#> Warning: Cannot create full citation for epidemiological parameters without bibliographic information
#> see ?as_epiparameter for help.
#> No adequate summary statistics available to calculate the parameters of the NA distribution
#> Unparameterised <epiparameter> object
marburg_gt_epiparameter
#> Disease: Marburg Virus Disease
#> Pathogen: Marburg virus
#> Epi Parameter: human delay generation time
#> Study: Ajelli (2012). "<title not available>." _<journal not available>_.
#> Distribution: NA
#> Mean: 9 [95% CI: 8.2, 10] (Days)
#> Median: NA [NA% CI: NA, NA] (Days)
#> Range: [c(NA, NA)] (Days)
marburg_gt_epiparameter$summary_stats
#> $mean
#> [1] 9
#>
#> $mean_ci_limits
#> [1] 8.2 10.0
#>
#> $mean_ci
#> [1] 95
#>
#> $sd
#> [1] 5.4
#>
#> $sd_ci_limits
#> [1] 3.9 8.6
#>
#> $sd_ci
#> [1] 95
#>
#> $median
#> [1] NA
#>
#> $median_ci_limits
#> [1] NA NA
#>
#> $median_ci
#> [1] NA
#>
#> $dispersion
#> [1] NA
#>
#> $dispersion_ci_limits
#> [1] NA NA
#>
#> $dispersion_ci
#> [1] NA
#>
#> $quantiles
#> [1] NA
#>
#> $range
#> [1] NA NAEntries with probability distributions
For this example we will load the Ebola epidemiological parameters from the {epireview} package (as there are no entries for Marburg that have parametric distributions).
ebola_data <- load_epidata("ebola")
#> ℹ ebola does not have any extracted outbreaks
#> information. Outbreaks will be set to NULL.
#> ✔ Data loaded for ebolaWe will again subset the data to just use the epidemiological parameter table, and select those rows containing a serial interval.
ebola_params <- ebola_data$params
ebola_si_rows <- ebola_params[
ebola_params$parameter_type_short == "serial_interval",
]
ebola_si_rows
#> # A tibble: 19 × 78
#> id parameter_data_id covidence_id pathogen parameter_type parameter_value
#> <chr> <chr> <int> <chr> <chr> <dbl>
#> 1 f49a9… 466f684ff8286fbd… 506 Ebola v… Human delay -… 12
#> 2 c1e68… cb37cc4599953d47… 1471 Ebola v… Human delay -… 19.4
#> 3 08e06… 20eb9e7d7714183c… 1876 Ebola v… Human delay -… 11
#> 4 5a250… 115c169147af31f7… 1891 Ebola v… Human delay -… 11.1
#> 5 54159… 6fca288e3bca7dc0… 3138 Ebola v… Human delay -… 16.1
#> 6 f044b… 89e334ec3622ed27… 3776 Ebola v… Human delay -… 14
#> 7 df908… e62da97ac8648211… 4966 Ebola v… Human delay -… 14.2
#> 8 df908… d46ff8b0c2ff67b7… 4966 Ebola v… Human delay -… 7.1
#> 9 1b9d9… abb8b6aabf43ac86… 5924 Ebola v… Human delay -… 13.7
#> 10 39354… 2b270d400af4fcce… 5939 Ebola v… Human delay -… NA
#> 11 39354… 8a18cde4823cf9f7… 5939 Ebola v… Human delay -… NA
#> 12 39354… 10f3384f1550a778… 5939 Ebola v… Human delay -… NA
#> 13 50dea… 631ec65830a82fbe… 6346 Ebola v… Human delay -… 15.3
#> 14 86e39… 5c8d68c39d1c3b98… 15896 Ebola v… Human delay -… 15.3
#> 15 40a29… 7f4ab651c48511df… 17077 Ebola v… Human delay -… 15.3
#> 16 b76dc… 0c3e02f80addfccc… 17730 Ebola v… Human delay -… 12
#> 17 b76dc… c2e0739d6bc652e9… 17730 Ebola v… Human delay -… 11.7
#> 18 74b62… e2a59f5aa40ddbdf… 18536 Ebola v… Human delay -… 12.3
#> 19 66e1b… 4da557e3c2c22a10… 19083 Ebola v… Human delay -… NA
#> # ℹ 72 more variables: exponent <dbl>, parameter_unit <chr>,
#> # parameter_lower_bound <dbl>, parameter_upper_bound <dbl>,
#> # parameter_value_type <chr>, parameter_uncertainty_single_value <dbl>,
#> # parameter_uncertainty_singe_type <chr>,
#> # parameter_uncertainty_lower_value <dbl>,
#> # parameter_uncertainty_upper_value <dbl>, parameter_uncertainty_type <chr>,
#> # cfr_ifr_numerator <int>, cfr_ifr_denominator <int>, …We will select an entry that has estimated and reported a Weibull distribution:
ebola_si <- ebola_si_rows[
ebola_si_rows$parameter_data_id == "0c3e02f80addfccc1017fa619fba76c5",
]
ebola_si
#> # A tibble: 1 × 78
#> id parameter_data_id covidence_id pathogen parameter_type parameter_value
#> <chr> <chr> <int> <chr> <chr> <dbl>
#> 1 b76dcc… 0c3e02f80addfccc… 17730 Ebola v… Human delay -… 12
#> # ℹ 72 more variables: exponent <dbl>, parameter_unit <chr>,
#> # parameter_lower_bound <dbl>, parameter_upper_bound <dbl>,
#> # parameter_value_type <chr>, parameter_uncertainty_single_value <dbl>,
#> # parameter_uncertainty_singe_type <chr>,
#> # parameter_uncertainty_lower_value <dbl>,
#> # parameter_uncertainty_upper_value <dbl>, parameter_uncertainty_type <chr>,
#> # cfr_ifr_numerator <int>, cfr_ifr_denominator <int>, …We can now convert this to an <epiparameter>
object.
ebola_si_epiparameter <- as_epiparameter(ebola_si)
#> Using Marziano (2023). "<title not available>." _<journal not available>_.
#> To retrieve the citation use the 'get_citation' function
#> Warning: Cannot create full citation for epidemiological parameters without bibliographic information
#> see ?as_epiparameter for help.
ebola_si_epiparameter
#> Disease: Ebola Virus Disease
#> Pathogen: Ebola virus
#> Epi Parameter: human delay serial interval
#> Study: Marziano (2023). "<title not available>." _<journal not available>_.
#> Distribution: weibull (Days)
#> Parameters:
#> shape: 1.760
#> scale: 10.140With the probability distribution for the serial interval we can
utilise some of the <epiparameter> methods. Here we
illustrate this by checking that the <epiparameter>
is parameterised, plotting the PDF and CDF, and generating 10 random
numbers sampling from the distribution.
is_parameterised(ebola_si_epiparameter)
#> [1] TRUE
plot(ebola_si_epiparameter)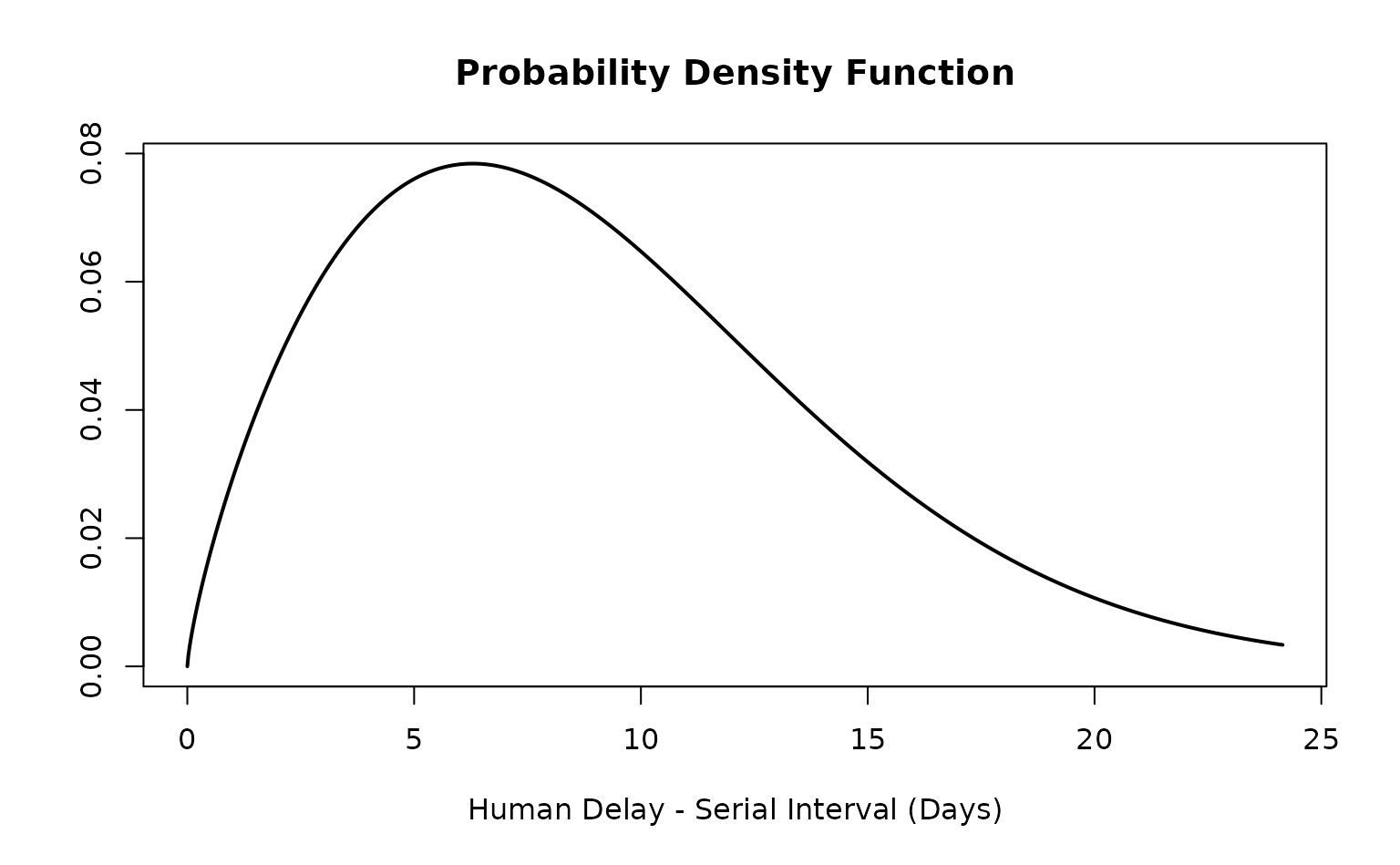
generate(ebola_si_epiparameter, times = 10)
#> [1] 8.1913357 17.4324678 1.4003823 6.3030243 0.9521153 7.7520633
#> [7] 4.8497066 7.3824799 4.6173973 6.4234107Specifying the probability distribution if unknown
There may be instances where a delay distribution is reported in the literature, but either a probability distribution is not fit to the data, or it is not reported which probability distribution the parameters correspond to. Therefore, there will not be a probability distribution specified in the {epireview} data. In these cases, and when a parametric probability distribution is required for a particular epidemiological task then assuming a probability distribution can be useful.
Please use this feature with caution. Assuming an
incorrect probability distribution and applying this in an
epidemiological method can lead to erroneous results. Additionally, if a
probability distribution is specified by the user it will overwrite any
probability distribution specified in input data (e.g. {epireview}
parameter data) which can lead to an error if the distribution name
supplied and parameters input are incompatible See
?as_epiparameter details for more information.
Just as the example above we will load the Ebola parameters using the
epireview::load_epidata() function and subset to just the
parameters ($params).
ebola_data <- load_epidata("ebola")
#> ℹ ebola does not have any extracted outbreaks
#> information. Outbreaks will be set to NULL.
#> ✔ Data loaded for ebola
ebola_params <- ebola_data$paramsHere we will use the serial interval for Ebola reported by Faye et al. (2015). This is stored, over two rows of the {epireview} parameter table, as the mean and standard deviation, but there is no probability distribution specified.
The code chunk below subsets the Ebola parameter table to just return the serial interval from Faye et al. (2015).
ebola_si <- ebola_params[
which(
grepl(pattern = "Faye", x = ebola_params$article_label, fixed = TRUE) &
grepl(pattern = "serial", ebola_params$parameter_type, fixed = TRUE)
),
]If we were to supply this data to as_epiparameter() we
would get an unparameterised <epiparameter> object
because no probability distribution is stated.
ebola_si_epiparameter <- as_epiparameter(ebola_si)
#> Using Faye (2015). "<title not available>." _<journal not available>_.
#> To retrieve the citation use the 'get_citation' function
#> Warning: Cannot create full citation for epidemiological parameters without bibliographic information
#> see ?as_epiparameter for help.
#> No adequate summary statistics available to calculate the parameters of the NA distribution
#> Unparameterised <epiparameter> object
ebola_si_epiparameter
#> Disease: Ebola Virus Disease
#> Pathogen: Ebola virus
#> Epi Parameter: human delay serial interval
#> Study: Faye (2015). "<title not available>." _<journal not available>_.
#> Distribution: NA
#> Mean: 14.2 [95% CI: 13.1, 15.5] (Days)
#> Median: NA [NA% CI: NA, NA] (Days)
#> Range: [c(NA, NA)] (Days)
is_parameterised(ebola_si_epiparameter)
#> [1] FALSEGiven that we can convert the mean and standard deviation into
parameters of a probability distribution if we assume a distribution
form, we can supply this data to as_epiparameter(). This
uses the parameter conversion functions in {epiparameter} (see
vignette("extract_convert", package = "epiparameter")).
ebola_si_epiparameter <- as_epiparameter(ebola_si, prob_distribution = "gamma")
#> Using Faye (2015). "<title not available>." _<journal not available>_.
#> To retrieve the citation use the 'get_citation' function
#> Warning: Cannot create full citation for epidemiological parameters without bibliographic information
#> see ?as_epiparameter for help.
#> Parameterising the probability distribution with the summary statistics.
#> Probability distribution is assumed not to be discretised or truncated.
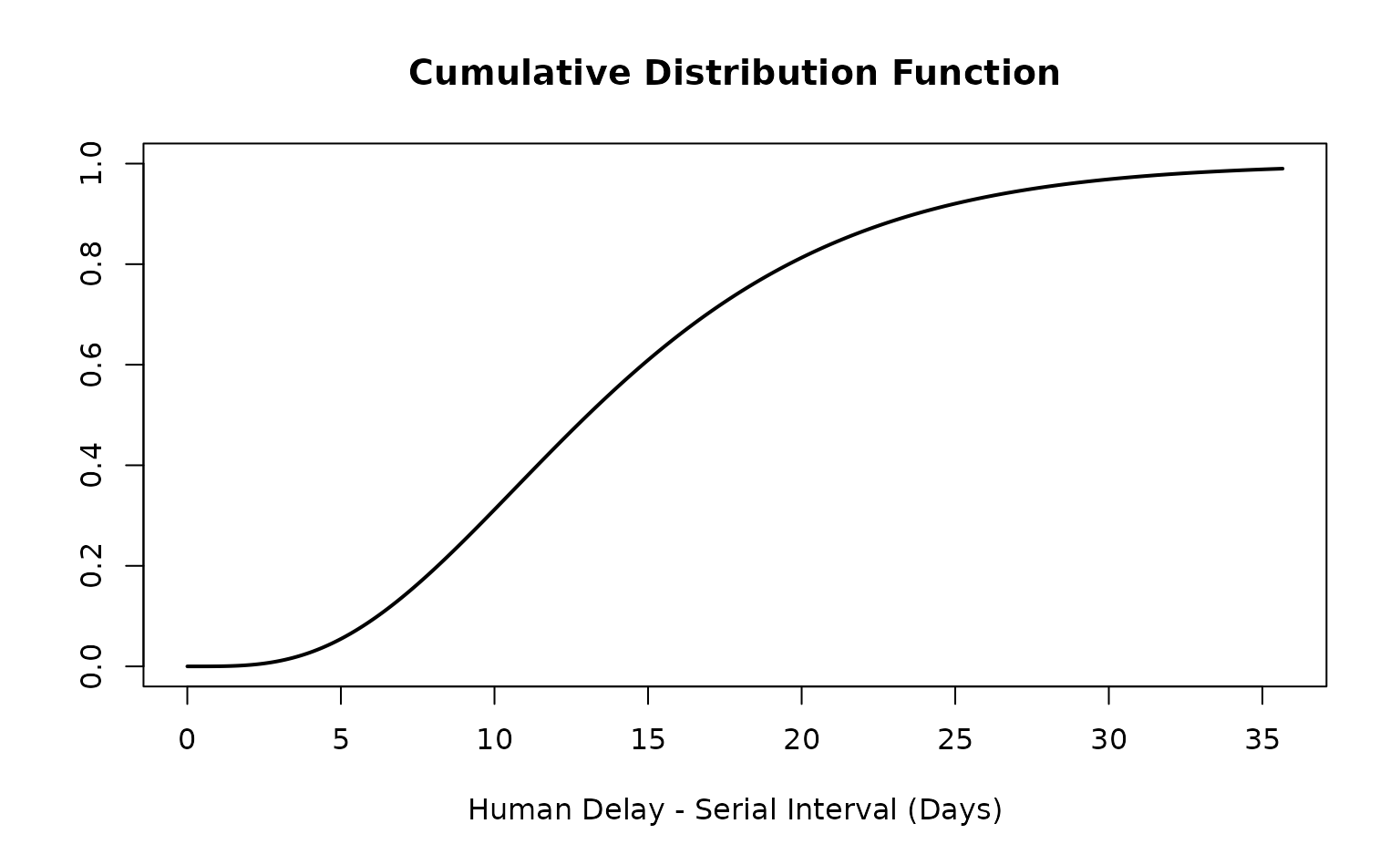
ebola_si_epiparameter
#> Disease: Ebola Virus Disease
#> Pathogen: Ebola virus
#> Epi Parameter: human delay serial interval
#> Study: Faye (2015). "<title not available>." _<journal not available>_.
#> Distribution: gamma (Days)
#> Parameters:
#> shape: 4.000
#> scale: 3.550
is_parameterised(ebola_si_epiparameter)
#> [1] TRUEThe Ebola serial interval <epiparameter> can now
be used for various probability distribution methods.
get_parameters(ebola_si_epiparameter)
#> shape scale
#> 4.00 3.55
density(ebola_si_epiparameter, at = 20)
#> [1] 0.03001206
plot(ebola_si_epiparameter)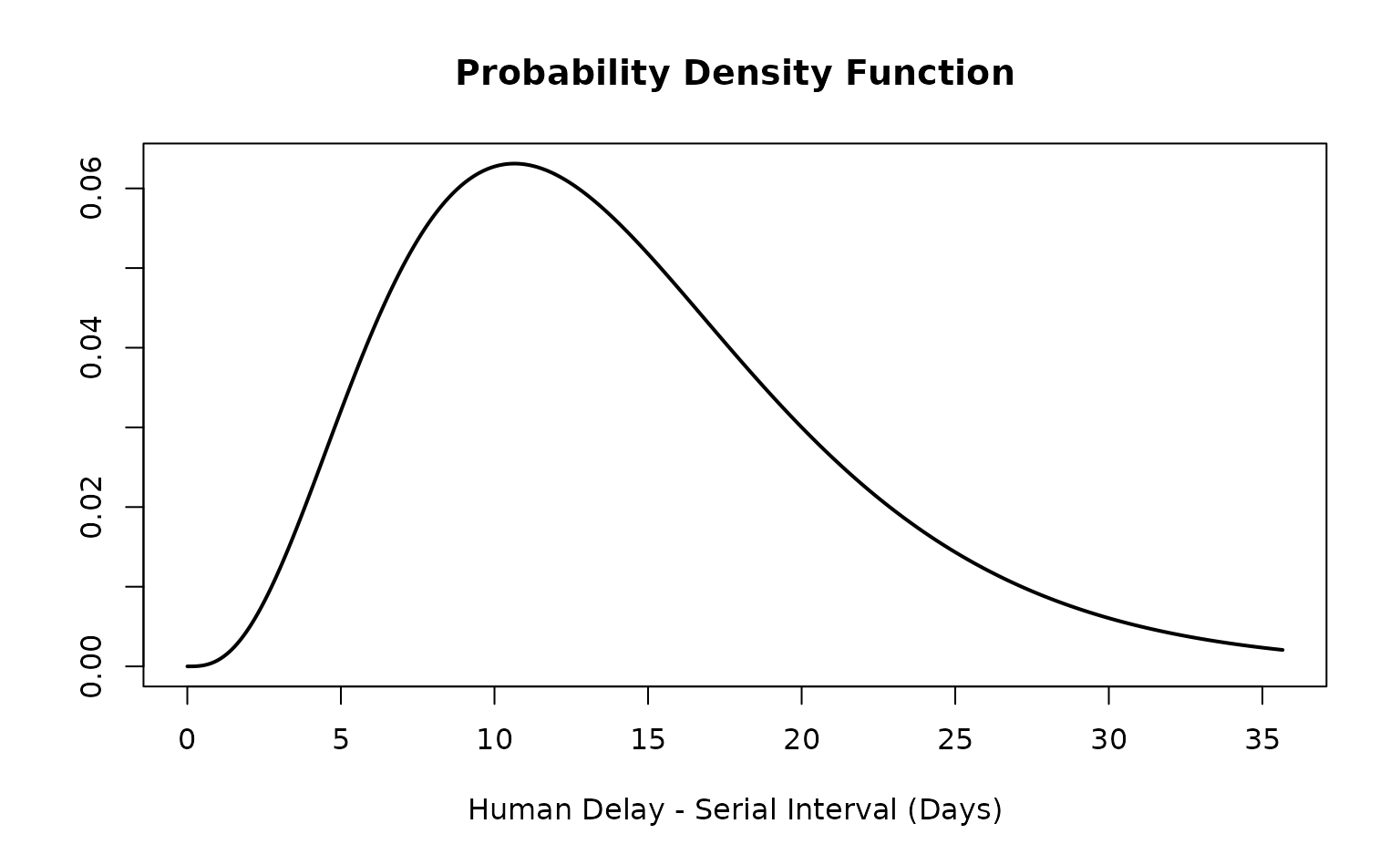
Limitations
- The database schema of {epireview} has evolved over time as the Imperial PERG team has extracted more pathogens.
- A list of parameter types is available in the {epireview} package
- It is important to differentiate between variability of the sample (e.g. sample standard deviation) and uncertainty of the estimate (e.g. 95% confidence interval or credible interval). From the database version of {epireview} of Zika, the PERG team will explicitly expose this to remove any ambiguity of the extracted data. Please note that in the Marburg, Lassa, and Ebola datasets, there may be some ambiguity between variability and uncertainty.
- This is functionality in {epiparameter} and {epireview} which will be developed and improved over the coming months.