It is often the case that in an infectious disease outbreak epidemiological parameters are required in order to characterise and model the dynamics of disease transmission and evaluate control strategies. In those scenarios, epidemiological parameters are commonly retrieved from the literature, and there is currently no library of parameters in order to contrast and compare different reported parameters for a range of infectious diseases and pathogens, from different published studies over time, of which some may be meta-analyses.
The {epiparameter} R package is a library of epidemiological parameters, with classes to handle this data and a set of functions to manipulate and use epidemiological parameters and distributions. The package also contains functionality for converting and extracting distribution parameters from summary statistics.
Use case
An outbreak of a known or potentially novel pathogen is detected and key parameters such as delay distributions (e.g. incubation period or serial interval) are required to interpret early data.
{epiparameter} can provide these distributions from a selection of published sources, such as past analysis of the same or similar pathogen, in order to provide relevant epidemiological parameters for new analysis.
This vignette will provide a introduction to the data stored within {epiparameter}, how to read it into R, manipulate the data, and the functions (and methods) implemented in the package to facilitate easy application of parameters into epidemiological pipelines.
Library of epidemiological parameters
First, we will introduce the library, or database, of epidemiological
parameters available from {epiparameter}. The library is stored
internally and can be read into R using the
epiparameter_db() function. By default all entries in the
library are returned.
db <- epiparameter_db()
#> Returning 125 results that match the criteria (100 are parameterised).
#> Use subset to filter by entry variables or single_epiparameter to return a single entry.
#> To retrieve the citation for each use the 'get_citation' function
db
#> # List of 125 <epiparameter> objects
#> Number of diseases: 23
#> ❯ Adenovirus ❯ Chikungunya ❯ COVID-19 ❯ Dengue ❯ Ebola Virus Disease ❯ Hantavirus Pulmonary Syndrome ❯ Human Coronavirus ❯ Influenza ❯ Japanese Encephalitis ❯ Marburg Virus Disease ❯ Measles ❯ MERS ❯ Mpox ❯ Parainfluenza ❯ Pneumonic Plague ❯ Rhinovirus ❯ Rift Valley Fever ❯ RSV ❯ SARS ❯ Smallpox ❯ West Nile Fever ❯ Yellow Fever ❯ Zika Virus Disease
#> Number of epi parameters: 13
#> ❯ case fatality risk ❯ generation time ❯ hospitalisation to death ❯ hospitalisation to discharge ❯ incubation period ❯ notification to death ❯ notification to discharge ❯ offspring distribution ❯ onset to death ❯ onset to discharge ❯ onset to hospitalisation ❯ onset to ventilation ❯ serial interval
#> [[1]]
#> Disease: Adenovirus
#> Pathogen: Adenovirus
#> Epi Parameter: incubation period
#> Study: Lessler J, Reich N, Brookmeyer R, Perl T, Nelson K, Cummings D (2009).
#> "Incubation periods of acute respiratory viral infections: a systematic
#> review." _The Lancet Infectious Diseases_.
#> doi:10.1016/S1473-3099(09)70069-6
#> <https://doi.org/10.1016/S1473-3099%2809%2970069-6>.
#> Distribution: lnorm (days)
#> Parameters:
#> meanlog: 1.723
#> sdlog: 0.231
#>
#> [[2]]
#> Disease: Human Coronavirus
#> Pathogen: Human_Cov
#> Epi Parameter: incubation period
#> Study: Lessler J, Reich N, Brookmeyer R, Perl T, Nelson K, Cummings D (2009).
#> "Incubation periods of acute respiratory viral infections: a systematic
#> review." _The Lancet Infectious Diseases_.
#> doi:10.1016/S1473-3099(09)70069-7
#> <https://doi.org/10.1016/S1473-3099%2809%2970069-7>.
#> Distribution: lnorm (days)
#> Parameters:
#> meanlog: 1.163
#> sdlog: 0.140
#>
#> [[3]]
#> Disease: SARS
#> Pathogen: SARS-Cov-1
#> Epi Parameter: incubation period
#> Study: Lessler J, Reich N, Brookmeyer R, Perl T, Nelson K, Cummings D (2009).
#> "Incubation periods of acute respiratory viral infections: a systematic
#> review." _The Lancet Infectious Diseases_.
#> doi:10.1016/S1473-3099(09)70069-8
#> <https://doi.org/10.1016/S1473-3099%2809%2970069-8>.
#> Distribution: lnorm (days)
#> Parameters:
#> meanlog: 1.386
#> sdlog: 0.593
#>
#> # ℹ 122 more elements
#> # ℹ Use `print(n = ...)` to see more elements.
#> # ℹ Use `parameter_tbl()` to see a summary table of the parameters.
#> # ℹ Explore database online at: https://epiverse-trace.github.io/epiparameter/articles/database.htmlThe output is a list of <epiparameter> objects,
where each element in the list corresponds to an entry in the parameter
database. To see a full list of the diseases and distributions stored in
the library use the parameter_tbl() function. Here we show
the first six rows of the output.
parameter_tbl(multi_epiparameter = db)
#> # Parameter table:
#> # A data frame: 125 × 7
#> disease pathogen epi_name prob_distribution author year sample_size
#> <chr> <chr> <chr> <chr> <chr> <dbl> <dbl>
#> 1 Adenovirus Adenovi… incubat… lnorm Lessl… 2009 14
#> 2 Human Coronavir… Human_C… incubat… lnorm Lessl… 2009 13
#> 3 SARS SARS-Co… incubat… lnorm Lessl… 2009 157
#> 4 Influenza Influen… incubat… lnorm Lessl… 2009 151
#> 5 Influenza Influen… incubat… lnorm Lessl… 2009 90
#> 6 Influenza Influen… incubat… lnorm Lessl… 2009 78
#> 7 Measles Measles… incubat… lnorm Lessl… 2009 55
#> 8 Parainfluenza Parainf… incubat… lnorm Lessl… 2009 11
#> 9 RSV RSV incubat… lnorm Lessl… 2009 24
#> 10 Rhinovirus Rhinovi… incubat… lnorm Lessl… 2009 28
#> # ℹ 115 more rowsparameter_tbl() can also subset the database supplied to
the function.
parameter_tbl(multi_epiparameter = db, disease = "Ebola")
#> # Parameter table:
#> # A data frame: 17 × 7
#> disease pathogen epi_name prob_distribution author year sample_size
#> <chr> <chr> <chr> <chr> <chr> <dbl> <dbl>
#> 1 Ebola Virus Dis… Ebola V… offspri… nbinom Lloyd… 2005 13
#> 2 Ebola Virus Dis… Ebola V… incubat… lnorm Eichn… 2011 196
#> 3 Ebola Virus Dis… Ebola V… onset t… gamma The E… 2018 14
#> 4 Ebola Virus Dis… Ebola V… incubat… gamma WHO E… 2015 1798
#> 5 Ebola Virus Dis… Ebola V… incubat… gamma WHO E… 2015 49
#> 6 Ebola Virus Dis… Ebola V… incubat… gamma WHO E… 2015 957
#> 7 Ebola Virus Dis… Ebola V… incubat… gamma WHO E… 2015 792
#> 8 Ebola Virus Dis… Ebola V… serial … gamma WHO E… 2015 305
#> 9 Ebola Virus Dis… Ebola V… serial … gamma WHO E… 2015 37
#> 10 Ebola Virus Dis… Ebola V… serial … gamma WHO E… 2015 147
#> 11 Ebola Virus Dis… Ebola V… serial … gamma WHO E… 2015 112
#> 12 Ebola Virus Dis… Ebola V… hospita… gamma WHO E… 2015 1167
#> 13 Ebola Virus Dis… Ebola V… hospita… gamma WHO E… 2015 1004
#> 14 Ebola Virus Dis… Ebola V… notific… gamma WHO E… 2015 2536
#> 15 Ebola Virus Dis… Ebola V… notific… gamma WHO E… 2015 1324
#> 16 Ebola Virus Dis… Ebola V… onset t… gamma WHO E… 2015 2741
#> 17 Ebola Virus Dis… Ebola V… onset t… gamma WHO E… 2015 1335More details on the data collation and the library of parameters can be found in the Data Collation and Synthesis Protocol vignette.
Single set of epidemiological parameters
{epiparameter} introduces a new class for working with
epidemiological parameters in R: <epiparameter>,
contains the name of the disease, the name of the epidemiological
distribution, parameters (if available) and citation information of
parameter source, as well as other information. This is the core data
structure in the {epiparameter} package and holds a single set of
epidemiological parameters.
An <epiparameter> object can be:
- Pulled from database (
epiparameter_db())
# <epiparameter> from database
# fetch <epiparameter> for COVID-19 incubation period from database
# return only a single <epiparameter>
covid_incubation <- epiparameter_db(
disease = "COVID-19",
epi_name = "incubation period",
single_epiparameter = TRUE
)
#> Using Linton N, Kobayashi T, Yang Y, Hayashi K, Akhmetzhanov A, Jung S, Yuan
#> B, Kinoshita R, Nishiura H (2020). "Incubation Period and Other
#> Epidemiological Characteristics of 2019 Novel Coronavirus Infections
#> with Right Truncation: A Statistical Analysis of Publicly Available
#> Case Data." _Journal of Clinical Medicine_. doi:10.3390/jcm9020538
#> <https://doi.org/10.3390/jcm9020538>..
#> To retrieve the citation use the 'get_citation' function
covid_incubation
#> Disease: COVID-19
#> Pathogen: SARS-CoV-2
#> Epi Parameter: incubation period
#> Study: Linton N, Kobayashi T, Yang Y, Hayashi K, Akhmetzhanov A, Jung S, Yuan
#> B, Kinoshita R, Nishiura H (2020). "Incubation Period and Other
#> Epidemiological Characteristics of 2019 Novel Coronavirus Infections
#> with Right Truncation: A Statistical Analysis of Publicly Available
#> Case Data." _Journal of Clinical Medicine_. doi:10.3390/jcm9020538
#> <https://doi.org/10.3390/jcm9020538>.
#> Distribution: lnorm (days)
#> Parameters:
#> meanlog: 1.525
#> sdlog: 0.629- Created manually (using the class constructor function:
epiparameter())
# <epiparameter> using constructor function
covid_incubation <- epiparameter(
disease = "COVID-19",
pathogen = "SARS-CoV-2",
epi_name = "incubation period",
prob_distribution = create_prob_distribution(
prob_distribution = "gamma",
prob_distribution_params = c(shape = 2, scale = 1)
),
summary_stats = create_summary_stats(mean = 2),
citation = create_citation(
author = person(
given = list("John", "Amy"),
family = list("Smith", "Jones")
),
year = 2022,
title = "COVID Incubation Period",
journal = "Epi Journal",
doi = "10.27861182.x"
)
)
#> Using Smith J, Jones A (2022). "COVID Incubation Period." _Epi Journal_.
#> doi:10.27861182.x <https://doi.org/10.27861182.x>.
#> To retrieve the citation use the 'get_citation' function
covid_incubation
#> Disease: COVID-19
#> Pathogen: SARS-CoV-2
#> Epi Parameter: incubation period
#> Study: Smith J, Jones A (2022). "COVID Incubation Period." _Epi Journal_.
#> doi:10.27861182.x <https://doi.org/10.27861182.x>.
#> Distribution: gamma (NA)
#> Parameters:
#> shape: 2.000
#> scale: 1.000Not all arguments are specified in the example using the class
constructor (epiparameter()) above, for example the
metadata or parameter uncertainty
(uncertainty) is not provided. See the help documentation
for the epiparameter() function using
?epiparameter to see each argument. Also see documentation
for <epiparameter> helper functions, e.g.,
?create_citation().
Manually creating <epiparameter> objects can be
especially useful if new parameter estimates become available but are
not yet incorporated into the {epiparameter} library.
As seen in the examples in this vignette, the
<epiparameter> class has a custom printing method
which shows the disease, pathogen (if known), the epidemiological
distribution, a citation of the study the parameters are from and the
probability distribution and parameter of that distribution (if
available).
Benefit of <epiparameter>
By providing a consistent and robust object to store epidemiological
parameters, <epiparameter> objects can be applied in
epidemiological pipelines, for example {episoap}. The data
contained within the object (e.g. parameter values, pathogen type, etc.)
can be modified but the pipeline will continue to operate because the
class is unchanged.
The probability distribution (prob_distribution)
argument requires the distribution specified in the standard R naming.
In some cases these are the same as the distribution’s name, e.g.,
gamma and weibull. Examples of where the
distribution name and R name differ are lognormal and
lnorm, negative binomial and nbinom, geometric
and geom, and poisson and pois.
Subsetting database
The database can be subset directly by
epiparameter_db(). Here the results can be subset by
author. It is recommended to use the family name of the first author
instead of the full name. Only the first author will be matched when the
entry is from a source with multiple authors.
epiparameter_db(
disease = "COVID-19",
epi_name = "incubation period",
author = "Linton"
)
#> Returning 3 results that match the criteria (3 are parameterised).
#> Use subset to filter by entry variables or single_epiparameter to return a single entry.
#> To retrieve the citation for each use the 'get_citation' function
#> # List of 3 <epiparameter> objects
#> Number of diseases: 1
#> ❯ COVID-19
#> Number of epi parameters: 1
#> ❯ incubation period
#> [[1]]
#> Disease: COVID-19
#> Pathogen: SARS-CoV-2
#> Epi Parameter: incubation period
#> Study: Linton N, Kobayashi T, Yang Y, Hayashi K, Akhmetzhanov A, Jung S, Yuan
#> B, Kinoshita R, Nishiura H (2020). "Incubation Period and Other
#> Epidemiological Characteristics of 2019 Novel Coronavirus Infections
#> with Right Truncation: A Statistical Analysis of Publicly Available
#> Case Data." _Journal of Clinical Medicine_. doi:10.3390/jcm9020538
#> <https://doi.org/10.3390/jcm9020538>.
#> Distribution: lnorm (days)
#> Parameters:
#> meanlog: 1.456
#> sdlog: 0.555
#>
#> [[2]]
#> Disease: COVID-19
#> Pathogen: SARS-CoV-2
#> Epi Parameter: incubation period
#> Study: Linton N, Kobayashi T, Yang Y, Hayashi K, Akhmetzhanov A, Jung S, Yuan
#> B, Kinoshita R, Nishiura H (2020). "Incubation Period and Other
#> Epidemiological Characteristics of 2019 Novel Coronavirus Infections
#> with Right Truncation: A Statistical Analysis of Publicly Available
#> Case Data." _Journal of Clinical Medicine_. doi:10.3390/jcm9020538
#> <https://doi.org/10.3390/jcm9020538>.
#> Distribution: lnorm (days)
#> Parameters:
#> meanlog: 1.611
#> sdlog: 0.472
#>
#> [[3]]
#> Disease: COVID-19
#> Pathogen: SARS-CoV-2
#> Epi Parameter: incubation period
#> Study: Linton N, Kobayashi T, Yang Y, Hayashi K, Akhmetzhanov A, Jung S, Yuan
#> B, Kinoshita R, Nishiura H (2020). "Incubation Period and Other
#> Epidemiological Characteristics of 2019 Novel Coronavirus Infections
#> with Right Truncation: A Statistical Analysis of Publicly Available
#> Case Data." _Journal of Clinical Medicine_. doi:10.3390/jcm9020538
#> <https://doi.org/10.3390/jcm9020538>.
#> Distribution: lnorm (days)
#> Parameters:
#> meanlog: 1.525
#> sdlog: 0.629
#>
#> # ℹ Use `parameter_tbl()` to see a summary table of the parameters.
#> # ℹ Explore database online at: https://epiverse-trace.github.io/epiparameter/articles/database.htmlThe results can be further subset using the subset
argument, for example subset = sample_size > 100 will
return entries with a sample size greater than 100. See
?epiparameter_db() for details on how to use this argument
to subset which database entries get returned.
If a single <epiparameter> is required then the
single_epiparameter argument can be set to
TRUE and this will return a single set of epidemiological
parameters (i.e. one delay distribution), if available. If multiple
entries in the parameter library match the search criteria (e.g. disease
type) then the entries that are parameterised (i.e. distribution
parameters are known), account for right truncation when inferred, and
were estimated from the largest sample size are preferentially selected
(in that order).
epiparameter_db(disease = "SARS", single_epiparameter = TRUE)
#> Using Lessler J, Reich N, Brookmeyer R, Perl T, Nelson K, Cummings D (2009).
#> "Incubation periods of acute respiratory viral infections: a systematic
#> review." _The Lancet Infectious Diseases_.
#> doi:10.1016/S1473-3099(09)70069-8
#> <https://doi.org/10.1016/S1473-3099%2809%2970069-8>..
#> To retrieve the citation use the 'get_citation' function
#> Disease: SARS
#> Pathogen: SARS-Cov-1
#> Epi Parameter: incubation period
#> Study: Lessler J, Reich N, Brookmeyer R, Perl T, Nelson K, Cummings D (2009).
#> "Incubation periods of acute respiratory viral infections: a systematic
#> review." _The Lancet Infectious Diseases_.
#> doi:10.1016/S1473-3099(09)70069-8
#> <https://doi.org/10.1016/S1473-3099%2809%2970069-8>.
#> Distribution: lnorm (days)
#> Parameters:
#> meanlog: 1.386
#> sdlog: 0.593Distribution functions
<epiparameter> objects store distributions, and
mathematical functions of these distribution can easily be extracted
directly from them. It is often useful to access the probability density
function, cumulative distribution function, quantiles of the
distribution, or generate random numbers from the distribution in the
<epiparameter> object. The distribution functions in
{epiparameter} allow users to easily use these.
ebola_incubation <- epiparameter_db(
disease = "Ebola",
epi_name = "incubation period",
single_epiparameter = TRUE
)
#> Using WHO Ebola Response Team, Agua-Agum J, Ariyarajah A, Aylward B, Blake I,
#> Brennan R, Cori A, Donnelly C, Dorigatti I, Dye C, Eckmanns T, Ferguson
#> N, Formenty P, Fraser C, Garcia E, Garske T, Hinsley W, Holmes D,
#> Hugonnet S, Iyengar S, Jombart T, Krishnan R, Meijers S, Mills H,
#> Mohamed Y, Nedjati-Gilani G, Newton E, Nouvellet P, Pelletier L,
#> Perkins D, Riley S, Sagrado M, Schnitzler J, Schumacher D, Shah A, Van
#> Kerkhove M, Varsaneux O, Kannangarage N (2015). "West African Ebola
#> Epidemic after One Year — Slowing but Not Yet under Control." _The New
#> England Journal of Medicine_. doi:10.1056/NEJMc1414992
#> <https://doi.org/10.1056/NEJMc1414992>..
#> To retrieve the citation use the 'get_citation' function
density(ebola_incubation, at = 0.5)
#> [1] 0.03608013
cdf(ebola_incubation, q = 0.5)
#> [1] 0.01178094
quantile(ebola_incubation, p = 0.5)
#> [1] 8.224347
generate(ebola_incubation, times = 10)
#> [1] 7.089471 25.516370 29.272107 12.667909 13.194056 5.102120 6.318984
#> [8] 11.372114 9.391204 9.740407Plotting epidemiological distributions
<epiparameter> objects can easily be plotted to
see the PDF and CDF of distribution.
plot(ebola_incubation)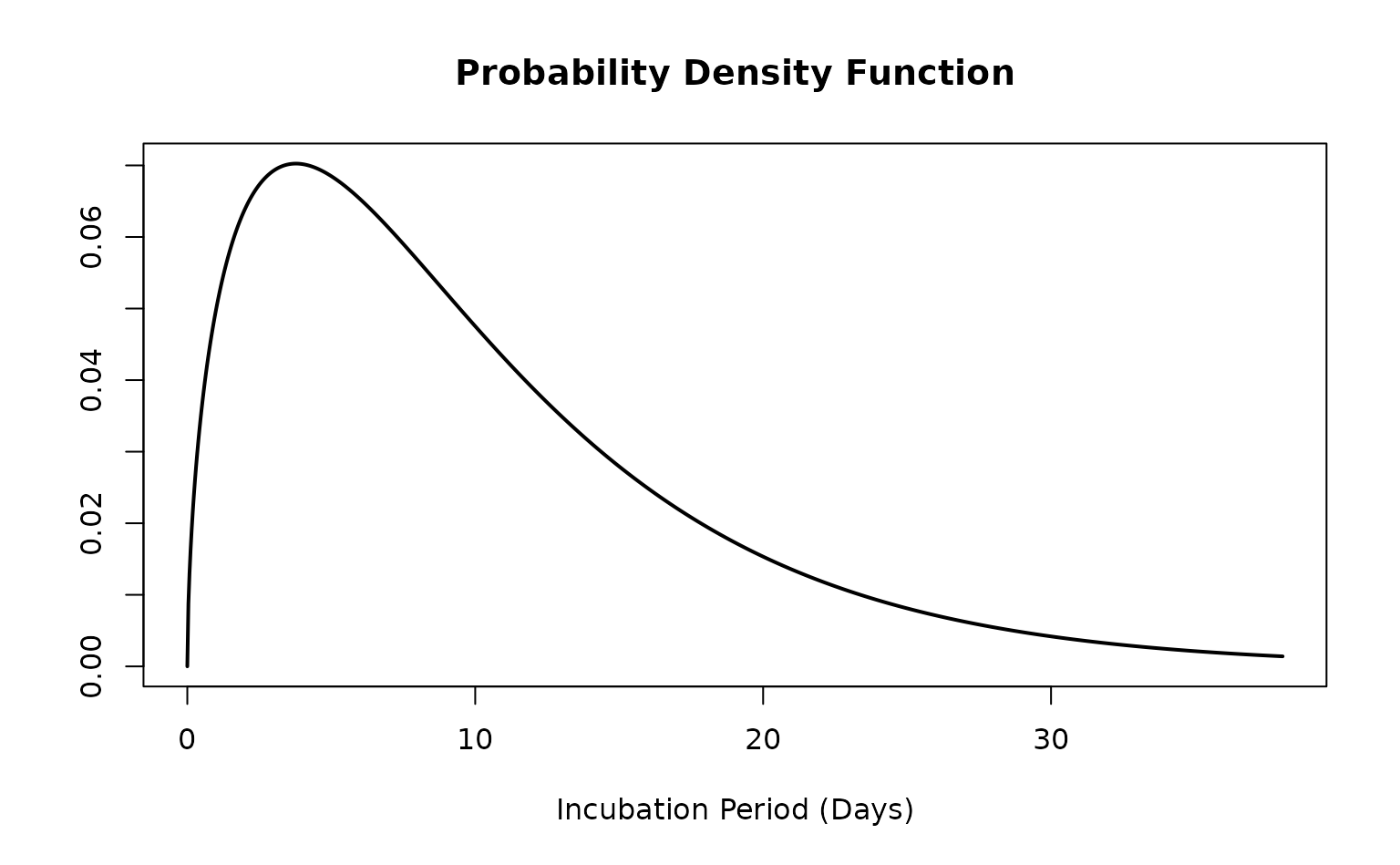
The default plotting range for time since infection is from zero to
the 99th quantile of the distribution. This can be altered by specifying
the xlim argument when plotting an
<epiparameter> object.
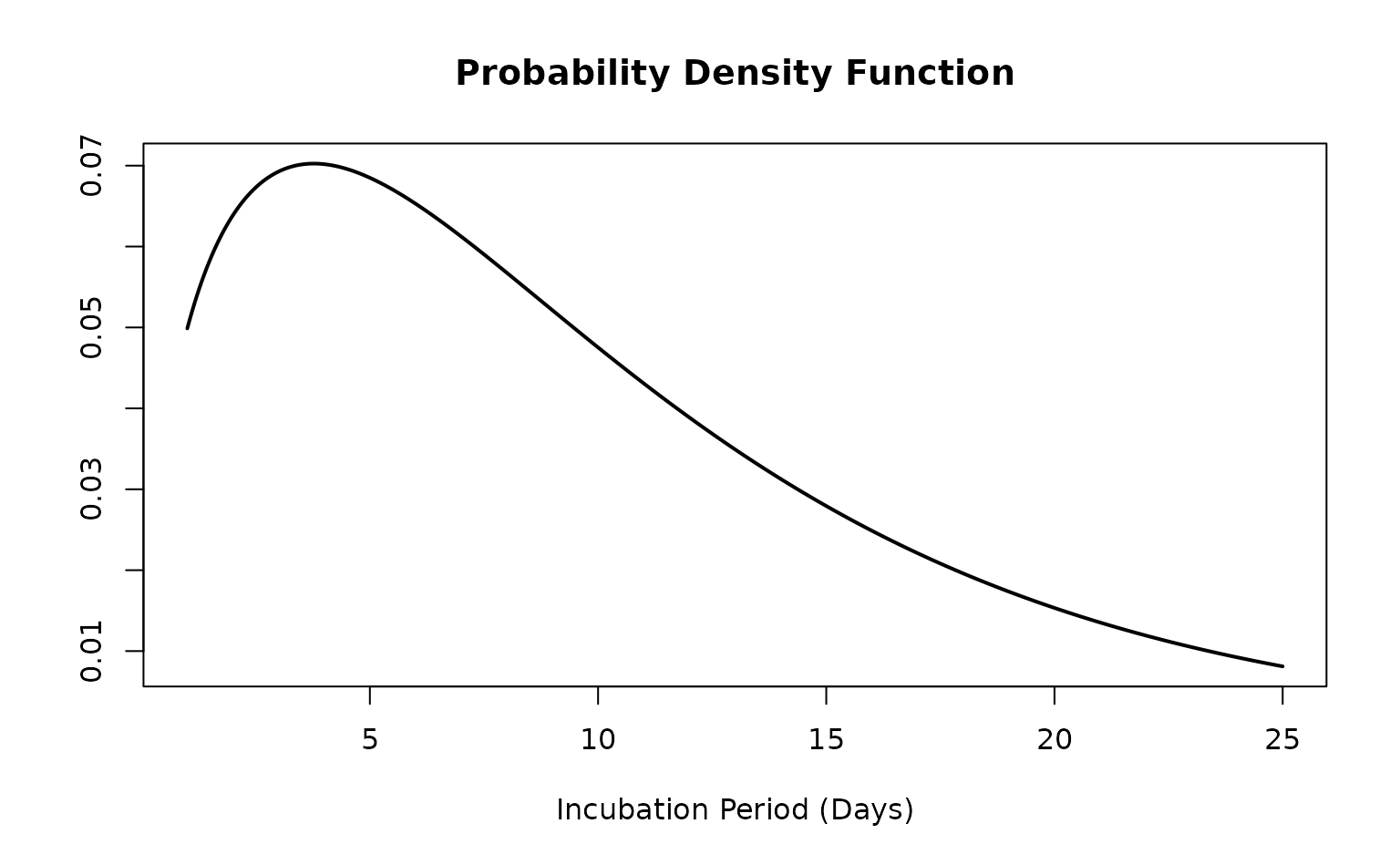
This plotting function can be useful for visually comparing
epidemiological distributions from different publications on the same
disease. In addition, plotting the distribution after manually creating
an <epiparameter> help to check that the parameters
are sensible and produce the expected distribution.
Accessors
The <epiparameter> class also has accessor
functions that can help access elements from the object in a
standardised format.
get_parameters(ebola_incubation)
#> shape scale
#> 1.577781 6.528155
get_citation(ebola_incubation)
#> WHO Ebola Response Team, Agua-Agum J, Ariyarajah A, Aylward B, Blake I,
#> Brennan R, Cori A, Donnelly C, Dorigatti I, Dye C, Eckmanns T, Ferguson
#> N, Formenty P, Fraser C, Garcia E, Garske T, Hinsley W, Holmes D,
#> Hugonnet S, Iyengar S, Jombart T, Krishnan R, Meijers S, Mills H,
#> Mohamed Y, Nedjati-Gilani G, Newton E, Nouvellet P, Pelletier L,
#> Perkins D, Riley S, Sagrado M, Schnitzler J, Schumacher D, Shah A, Van
#> Kerkhove M, Varsaneux O, Kannangarage N (2015). "West African Ebola
#> Epidemic after One Year — Slowing but Not Yet under Control." _The New
#> England Journal of Medicine_. doi:10.1056/NEJMc1414992
#> <https://doi.org/10.1056/NEJMc1414992>.Parameter conversion and extraction
Conversion
Parameters are often reported in the literature as mean and standard
deviation (or variance). These summary statistics can often be
(analytically) converted to the parameters of the distribution using the
conversion function in the package
(convert_summary_stats_to_params()). We also provide
conversion functions in the opposite direction, parameters to summary
statistics (convert_params_to_summary_stats()).
Extraction
The functions extract_param() handles all the extraction
of parameter estimates from summary statistics. The two extractions
currently supported in {epiparameter} are from percentiles and from
median and range.
Adding library entries and contributing to {epiparameter}
If a set of epidemiological parameter has been inferred and is known to the user but has not yet been incorporated into the {epiparameter} database, these parameters can be manually added to the library.
# wrap <epiparameter> in list to append to database
new_db <- append(db, covid_incubation)
tail(new_db, n = 3)
#> [[1]]
#> Disease: Chikungunya
#> Pathogen: Chikungunya Virus
#> Epi Parameter: generation time
#> Study: Guzzetta G, Vairo F, Mammone A, Lanini S, Poletti P, Manica M, Rosa R,
#> Caputo B, Solimini A, della Torre A, Scognamiglio P, Zumla A, Ippolito
#> G, Merler S (2020). "Spatial modes for transmission of chikungunya
#> virus during a large chikungunya outbreak in Italy: a modeling
#> analysis." _BMC Medicine_. doi:10.1186/s12916-020-01674-y
#> <https://doi.org/10.1186/s12916-020-01674-y>.
#> Distribution: gamma (days)
#> Parameters:
#> shape: 8.633
#> scale: 1.447
#>
#> [[2]]
#> Disease: Chikungunya
#> Pathogen: Chikungunya Virus
#> Epi Parameter: case fatality risk
#> Study: de Souza W, de Lima S, Mello L, Candido D, Buss L, Whittaker C, Claro
#> I, Chandradeva N, Granja F, de Jesus R, Lemos P, Toledo-Teixeira D,
#> Barbosa P, Firmino A, Amorim M, Duarte L, Pessoa Jr I, Forato J,
#> Vasconcelos I, Maximo A, Araújo E, Mello L, Sabino E, Proença-Módena J,
#> Faria N, Weaver S (2023). "Spatiotemporal dynamics and recurrence of
#> chikungunya virus in Brazil: an epidemiological study." _The Lancet
#> Microbe_. doi:10.1016/S2666-5247(23)00033-2
#> <https://doi.org/10.1016/S2666-5247%2823%2900033-2>.
#> Parameters: <no parameters>
#> Mean: 1.3 (deaths per 1000 cases)
#>
#> [[3]]
#> Disease: COVID-19
#> Pathogen: SARS-CoV-2
#> Epi Parameter: incubation period
#> Study: Smith J, Jones A (2022). "COVID Incubation Period." _Epi Journal_.
#> doi:10.27861182.x <https://doi.org/10.27861182.x>.
#> Distribution: gamma (NA)
#> Parameters:
#> shape: 2.000
#> scale: 1.000Note that this only adds the parameters to the library in the environment, and does not save to the database file in the package. Hence, if you restart your R session, you will lose the changes.
The library of epidemiological parameters is a living database, so as new studies are published we hope to incorporate these. Searching for and recording parameters in the database is extremely time-consuming, so we welcome contributions of new parameters by either making a pull request to the package or adding information to the contributing spreadsheet. These will be incorporated into the database by the package maintainers and your contributions will be acknowledged. See the Data Collation and Synthesis Protocol vignette on information about contributing to the library of epidemiological parameters.
