Content from Read case data
Last updated on 2026-02-24 | Edit this page
Overview
Questions
- Where do you usually store outbreak data?
- What data formats do you commonly use for analysis?
- Can you import data directly from servers and health information systems?
Objectives
- Identify common sources of outbreak data.
- Import outbreak data from multiple formats into
Renvironment. - Access and retrieve data from remote servers and health information systems using APIs.
Prerequisites
This episode requires you to be familiar with Data science: Basic tasks with R.
Introduction
The first step in outbreak analysis is importing your dataset into
the R environment. Data can come from local sources, like
files on your computer, or external sources, like databases and health
information systems (HIS).
Outbreak data takes many forms. It may be sorted as a flat file in various formats, housed in relational database management systems (RDBMS), or managed through specialized HIS like SORMAS and DHIS2. These HISs offer application programming interfaces (APIs) that allow authorized users to modify and retrieve data entries efficiently, making them particularly valuable for large-scale institutional health data collection and storage.
This episode demonstrates how to read case data from each of these
sources. Let’s begin by loading the packages we’ll need. We will use
rio to read data stored in files and
readepi to access data from RDBMS and HIS. We will also
load here to locate file paths within your project
directory, and tidyverse, which includes
magrittr (providing the pipe operator
%>%) and dplyr (for data manipulation).
The pipe operator allows us to chain functions together seamlessly.
R
# Load packages
library(tidyverse) # for {dplyr} functions and the pipe %>%
library(rio) # for importing data from files
library(here) # for easy file referencing
library(readepi) # for importing data directly from RDBMS or HIS
library(dbplyr) # for a database backend for {dplyr}
The double-colon (::)
operator
The double-colon :: in R lets you call a
specific function from a package without loading the entire package. For
example, dplyr::filter(data, condition) uses the
filter() function from the dplyr package,
without requiring library(dplyr).
This notation serves two purposes: it makes code more readable by explicitly showing which package each function comes from, and it prevents namespace conflicts that occur when multiple packages contain functions with the same name.
Setup a project and folder
- Create an RStudio project. If needed, follow this how-to guide on “Hello RStudio Projects” to create one.
- Inside the RStudio project, create a
data/folder. - Download ebola_cases_2.csv
and marburg.zip
CSV files, and save them inside the
data/folder.
Reading from files
Several packages are available for importing outbreak data stored in
individual files into R. These include {rio}, {readr} from the
tidyverse, {io}, {ImportExport},
and {data.table}.
Together, these packages offer methods to read single or multiple files
in a wide range of formats.
The below example shows how to import a csv file into
R environment using the rio package. We use
the here package to tell R to look for the file in the
data/ folder of your project, and
dplyr::as_tibble() to convert into a tidier format for
subsequent analysis in R.
R
# read data
# e.g., if the path to our file is "data/raw-data/ebola_cases_2.csv" then:
ebola_confirmed <- rio::import(
here::here("data", "raw-data", "ebola_cases_2.csv")
) %>%
dplyr::as_tibble() # for a simple data frame output
# preview data
ebola_confirmed
OUTPUT
# A tibble: 120 × 4
year month day confirm
<int> <int> <int> <int>
1 2014 5 18 1
2 2014 5 20 2
3 2014 5 21 4
4 2014 5 22 6
5 2014 5 23 1
6 2014 5 24 2
7 2014 5 26 10
8 2014 5 27 8
9 2014 5 28 2
10 2014 5 29 12
# ℹ 110 more rowsYou can use the same approach to import other file formats such as
tsv, xlsx, and more.
Why should we use the {here} package?
The here package is designed to simplify file referencing in R projects by providing a reliable way to construct file paths relative to the project root. The main reason to use is for cross-environment compatibility.
It works across different operating systems (Windows, Mac, Linux) without needing to adjust file paths.
- On Windows, paths are written using backslashes (
\) as the separator between folder names:"data\raw-data\file.csv". - On Unix based operating systems such as macOS or Linux the forward
slash (
/) is used as the path separator:"data/raw-data/file.csv".
The here package reinforces the reproducibility of your work across multiple operating systems. If you are interested in reproducibility, we invite you to read this tutorial to increase the openess, sustainability, and reproducibility of your epidemic analysis with R
Reading compressed data
Can you read data from a compressed file in R?
Download this zip
file containing Marburg outbreak data and then import it to your
R environment.
You can check the full list of supported file formats in the rio package on the package website. To see the list of supported formats in rio, run:
R
rio::install_formats()
R
rio::import(here::here("data", "Marburg.zip"))
Reading from databases
The readepi library contains functions that allow you
to import data directly from RDBMS. The
readepi::read_rdbms() function supports importing data from
servers such as Microsoft SQL, MySQL, PostgreSQL, and SQLite. It build
on the {DBI} package, which
provides a general interface for interacting RDBMS.
Advantages of reading data directly from a database?
Importing data directly from a database optimizes the memory usage in the R session. By processing the database with “queries” (e.g., SELECT, FILTER, GROUP BY) before extraction, you reduce the memory load in our RStudio session. In contrast, loading an entire dataset into R for manipulation can consume more RAM than your local machine can handle, potentially causing RStudio to slow down or freeze.
RDBMS also enable multiple users to access, store, and analyze parts of the dataset simultaneously without transferring individual files. This eliminates the version control problems that arise when multiple file copies circulate among users.
1. Connect with a database
You can use the readepi::login() function to establish a
connection to the database, as shown below:
R
# establish the connection to a test MySQL database
rdbms_login <- readepi::login(
from = "mysql-rfam-public.ebi.ac.uk",
type = "MySQL",
user_name = "rfamro",
password = "",
driver_name = "",
db_name = "Rfam",
port = 4497
)
OUTPUT
✔ Logged in successfully!R
rdbms_login
OUTPUT
<Pool> of MySQLConnection objects
Objects checked out: 0
Available in pool: 1
Max size: Inf
Valid: TRUEThe function parameters are:
-
from: The database server address (mysql-rfam-public.ebi.ac.uk) -
type: The type of database system (“MySQL”) -
user_name: The username for authentication (“rfamro”) -
password: The password (empty string “” indicates no password required for this public test database) -
driver_name: The database driver (empty string uses the default driver) -
db_name: The specific database to connect to (“Rfam”) -
port: The port number for the connection (4497)
The function returns a connection object stored in variable
rdbms_login, which can then be used to query and retrieve
data from the database.
Callout
Note: This example uses a public test database from the European Bioinformatics Institute, which is why no password is required. Access to it may be limited by organizational network restrictions, but it should work normally on home networks.
2. Access the list of tables from the database
The readepi::show_tables() function retrieves the full
list of table names from a database:
R
# get the table names
tables <- readepi::show_tables(login = rdbms_login)
tables
In a relational database, you typically have multiple tables. Each table represents a specific entity (e.g., patients, care units, treatments). Tables are linked through common identifiers called primary keys or foreign keys.
3. Read data from a table in a database
Use the readepi::read_rdbms() function to import data
from a database table. It accepts either an SQL query or a list of query
parameters, as demonstrated in the code chunk below.
R
# import data from the 'author' table using an SQL query
dat <- readepi::read_rdbms(
login = rdbms_login,
query = "select * from author"
)
# import data from the 'author' table using a list of parameters
dat <- readepi::read_rdbms(
login = rdbms_login,
query = list(table = "author", fields = NULL, filter = NULL)
)
Alternatively, you can read the data from the author
table using dplyr::tbl().
R
# import data from the 'author' table using an SQL query
dat <- rdbms_login %>%
dplyr::tbl(from = "author") %>%
dplyr::filter(initials == "A") %>%
dplyr::arrange(desc(author_id))
dat
OUTPUT
# Source: SQL [?? x 6]
# Database: mysql 8.0.32-24 [@mysql-rfam-public.ebi.ac.uk:/Rfam]
# Ordered by: desc(author_id)
author_id name last_name initials orcid synonyms
<int> <chr> <chr> <chr> <chr> <chr>
1 46 Roth A Roth A "" ""
2 42 Nahvi A Nahvi A "" ""
3 32 Machado Lima A Machado Lima A "" ""
4 31 Levy A Levy A "" ""
5 27 Gruber A Gruber A "0000-0003-1219-4239" ""
6 13 Chen A Chen A "" ""
7 6 Bateman A Bateman A "0000-0002-6982-4660" "" When you apply dplyr verbs to this database table, they are automatically translated into SQL queries:
R
# Show the SQL queries translated
dat %>%
dplyr::show_query()
OUTPUT
<SQL>
SELECT `author`.*
FROM `author`
WHERE (`initials` = 'A')
ORDER BY `author_id` DESC4. Extract data from the database
Use dplyr::collect() to force computation of a database
query and extract the output to your local computer.
R
# Pull all data down to a local tibble
dat %>%
dplyr::collect()
OUTPUT
# A tibble: 7 × 6
author_id name last_name initials orcid synonyms
<int> <chr> <chr> <chr> <chr> <chr>
1 46 Roth A Roth A "" ""
2 42 Nahvi A Nahvi A "" ""
3 32 Machado Lima A Machado Lima A "" ""
4 31 Levy A Levy A "" ""
5 27 Gruber A Gruber A "0000-0003-1219-4239" ""
6 13 Chen A Chen A "" ""
7 6 Bateman A Bateman A "0000-0002-6982-4660" "" Ideally, after specifying a set of queries, we can reduce the size of the input dataset to use in the environment of our R session.
Run SQL queries in R using {dbplyr}
Practice how to make relational database SQL queries using multiple
dplyr verbs like dplyr::left_join() among
tables before pulling out data to your local session with
dplyr::collect()!
You can also review the dbplyr R package. But for a step-by-step tutorial about SQL, we recommend you this tutorial about data management with SQL for Ecologist.
R
# SELECT FEW COLUMNS FROM ONE TABLE AND LEFT JOIN WITH ANOTHER TABLE
author <- rdbms_login %>%
dplyr::tbl(from = "author") %>%
dplyr::select(author_id, name)
family_author <- rdbms_login %>%
dplyr::tbl(from = "family_author") %>%
dplyr::select(author_id, rfam_acc)
dplyr::left_join(author, family_author, keep = TRUE) %>%
dplyr::show_query()
OUTPUT
Joining with `by = join_by(author_id)`OUTPUT
<SQL>
SELECT
`author`.`author_id` AS `author_id.x`,
`name`,
`family_author`.`author_id` AS `author_id.y`,
`rfam_acc`
FROM `author`
LEFT JOIN `family_author`
ON (`author`.`author_id` = `family_author`.`author_id`)R
dplyr::left_join(author, family_author, keep = TRUE) %>%
dplyr::collect()
OUTPUT
Joining with `by = join_by(author_id)`OUTPUT
# A tibble: 5,029 × 4
author_id.x name author_id.y rfam_acc
<int> <chr> <int> <chr>
1 1 Ames T 1 RF01831
2 2 Argasinska J 2 RF02554
3 2 Argasinska J 2 RF02555
4 2 Argasinska J 2 RF02722
5 2 Argasinska J 2 RF02720
6 2 Argasinska J 2 RF02719
7 2 Argasinska J 2 RF02721
8 2 Argasinska J 2 RF02670
9 2 Argasinska J 2 RF02718
10 2 Argasinska J 2 RF02668
# ℹ 5,019 more rowsReading from HIS APIs
Health data is increasingly stored in specialized HIS such as Fingertips, GoData, REDCap, DHIS2, SORMAS, etc. The current version of the readepi library allows importing data from DHIS2 and SORMAS. The subsections below demonstrate how to import data from these two systems.
Importing data from DHIS2
DHIS2 (District Health
Information System) is an open-source software that has revolutionized
global health information management. The
readepi::read_dhis2() function imports data from the DHIS2
Tracker system via
its API.
To successfully import data from DHIS2, you need to:
- Connect to the system using the
readepi::login()function - Provide the name or ID of the target program and organization unit
You can retrieve the IDs and names of available programs and
organization units using the get_programs() and
get_organisation_units() functions, respectively.
R
# establish the connection to the system
dhis2_login <- readepi::login(
type = "dhis2",
from = "https://smc.moh.gm/dhis",
user_name = "test",
password = "Gambia@123"
)
# get the names and IDs of the programs
programs <- readepi::get_programs(login = dhis2_login)
tibble::as_tibble(programs)
OUTPUT
# A tibble: 2 × 3
displayName id type
<chr> <chr> <chr>
1 "Child Registration & Treatment " E5IUQuHg3Mg tracker
2 "Daily Drug Reconciliations" I3bZrR6fLt8 aggregateR
# get the names and IDs of the organisation units
org_units <- readepi::get_organisation_units(login = dhis2_login)
tibble::as_tibble(org_units)
OUTPUT
# A tibble: 872 × 10
National_name National_id Regional_name Regional_id District_name District_id
<chr> <chr> <chr> <chr> <chr> <chr>
1 Gambia jvQPTsCLwPh Central Rive… gsMpbz5DQsM "Upper fulla… srjR5LWAoBD
2 Gambia jvQPTsCLwPh Western Regi… D18zNdCbRfO "Foni Jarrol… kAxFyJFfYV8
3 Gambia jvQPTsCLwPh Upper River … SHRxQEqOPJa "Sandu" iZQOFwckdXL
4 Gambia jvQPTsCLwPh Central Rive… gsMpbz5DQsM "Upper fulla… srjR5LWAoBD
5 Gambia jvQPTsCLwPh Upper River … SHRxQEqOPJa "Basse (Full… Ug7sj97icMt
6 Gambia jvQPTsCLwPh Upper River … SHRxQEqOPJa "Basse (Full… Ug7sj97icMt
7 Gambia jvQPTsCLwPh Central Rive… gsMpbz5DQsM "Sami" ZZNUH1LhS7k
8 Gambia jvQPTsCLwPh Western Regi… D18zNdCbRfO "Foni Jarrol… kAxFyJFfYV8
9 Gambia jvQPTsCLwPh Central Rive… gsMpbz5DQsM "Niamina Dan… T55lst07vTj
10 Gambia jvQPTsCLwPh Upper River … SHRxQEqOPJa "Tumana" xGYsUdiJb4L
# ℹ 862 more rows
# ℹ 4 more variables: `Operational Zone_name` <chr>,
# `Operational Zone_id` <chr>, `Town/Village_name` <chr>,
# `Town/Village_id` <chr>After retrieving organization units and program names from the DHIS2 database, we can import data using either names or coded IDs, as demonstrated in the code chunk below
R
data <- readepi::read_dhis2(
login = dhis2_login,
org_unit = "Keneba",
program = "Child Registration & Treatment "
)
tibble::as_tibble(data)
OUTPUT
# A tibble: 1,116 × 69
event tracked_entity org_unit ` SMC-CR Scan QR Code` SMC-CR Did the child…¹
<chr> <chr> <chr> <chr> <chr>
1 bgSDQb… yv7MOkGD23q Keneba SMC23-0510989 1
2 y4MKmP… nibnZ8h0Nse Keneba SMC2021-018089 1
3 yK7VG3… nibnZ8h0Nse Keneba SMC2021-018089 1
4 EmNflz… nibnZ8h0Nse Keneba SMC2021-018089 1
5 UF96ms… nibnZ8h0Nse Keneba SMC2021-018089 1
6 guQTwc… FomREQ2it4n Keneba SMC23-0510012 1
7 jbkRkL… FomREQ2it4n Keneba SMC23-0510012 1
8 AEeype… FomREQ2it4n Keneba SMC23-0510012 1
9 R30SPs… E5oAWGcdFT4 Keneba koika-smc-22897 1
10 nr03Qy… E5oAWGcdFT4 Keneba koika-smc-22897 1
# ℹ 1,106 more rows
# ℹ abbreviated name: ¹`SMC-CR Did the child previously received a card?`
# ℹ 64 more variables: `SMC-CR Child First Name1` <chr>,
# `SMC-CR Child Last Name` <chr>, `SMC-CR Date of Birth` <chr>,
# `SMC-CR Select Age Category ` <chr>, `SMC-CR Child gender1` <chr>,
# `SMC-CR Mother/Person responsible full name` <chr>,
# `SMC-CR Mother/Person responsible phone number1` <chr>, …R
# import data from DHIS2 using names
# import data from DHIS2 using IDs
data <- readepi::read_dhis2(
login = dhis2_login,
org_unit = "GcLhRNAFppR",
program = "E5IUQuHg3Mg"
)
tibble::as_tibble(data)
OUTPUT
# A tibble: 1,116 × 69
event tracked_entity org_unit ` SMC-CR Scan QR Code` SMC-CR Did the child…¹
<chr> <chr> <chr> <chr> <chr>
1 bgSDQb… yv7MOkGD23q Keneba SMC23-0510989 1
2 y4MKmP… nibnZ8h0Nse Keneba SMC2021-018089 1
3 yK7VG3… nibnZ8h0Nse Keneba SMC2021-018089 1
4 EmNflz… nibnZ8h0Nse Keneba SMC2021-018089 1
5 UF96ms… nibnZ8h0Nse Keneba SMC2021-018089 1
6 guQTwc… FomREQ2it4n Keneba SMC23-0510012 1
7 jbkRkL… FomREQ2it4n Keneba SMC23-0510012 1
8 AEeype… FomREQ2it4n Keneba SMC23-0510012 1
9 R30SPs… E5oAWGcdFT4 Keneba koika-smc-22897 1
10 nr03Qy… E5oAWGcdFT4 Keneba koika-smc-22897 1
# ℹ 1,106 more rows
# ℹ abbreviated name: ¹`SMC-CR Did the child previously received a card?`
# ℹ 64 more variables: `SMC-CR Child First Name1` <chr>,
# `SMC-CR Child Last Name` <chr>, `SMC-CR Date of Birth` <chr>,
# `SMC-CR Select Age Category ` <chr>, `SMC-CR Child gender1` <chr>,
# `SMC-CR Mother/Person responsible full name` <chr>,
# `SMC-CR Mother/Person responsible phone number1` <chr>, …Note that not all organization units are registered for a specific
program. To find which organization units are running a particular
program, use the get_program_org_units() function as shown
below:
R
# get the list of organisation units that run the program "E5IUQuHg3Mg"
target_org_units <- readepi::get_program_org_units(
login = dhis2_login,
program = "E5IUQuHg3Mg",
org_units = org_units
)
tibble::as_tibble(target_org_units)
OUTPUT
# A tibble: 26 × 3
org_unit_ids levels org_unit_names
<chr> <chr> <chr>
1 UrLrbEiWk3J Town/Village_name Sare Sibo
2 wlVsFVeHSTx Town/Village_name Jawo Kunda
3 kp0ZYUEqJE8 Town/Village_name Chewal
4 Wr3htgGxhBv Town/Village_name Madinayel
5 psyHoqeN2Tw Town/Village_name Bolibanna
6 MGBYonFM4y3 Town/Village_name Sare Mala
7 GcLhRNAFppR Town/Village_name Keneba
8 y1Z3KuvQyhI Town/Village_name Brikama
9 W3vH9yBUSei Town/Village_name Gidda
10 ISbNWYieHY8 Town/Village_name Song Kunda
# ℹ 16 more rowsCallout
Note: This example uses a DHIS2 system provided by the Ministry of Health of The Gambia for testing and development purposes. In practice, you should customize the parameters for your own DHIS2 instance.
Reading from Demo DHIS2 sever
The DHIS2 organization provides demo servers for development and testing. One of these is called stable-242-4, available at the link (“https://play.im.dhis2.org/stable-2-42-4”), and accessible with username “admin” and password “district”. Log into this server, list all available programs and organization units, and read data from one of these programs.
R
# establish the connection to the system
demo_login <- readepi::login(
type = "dhis2",
from = "https://play.im.dhis2.org/stable-2-42-4",
user_name = "admin",
password = "district"
)
# get the names and IDs of the programs
demo_programs <- readepi::get_programs(login = demo_login)
tibble::as_tibble(demo_programs)
OUTPUT
# A tibble: 14 × 3
displayName id type
<chr> <chr> <chr>
1 Antenatal care visit lxAQ7Zs9VYR aggregate
2 Child Programme IpHINAT79UW tracker
3 Contraceptives Voucher Program kla3mAPgvCH aggregate
4 Information Campaign q04UBOqq3rp aggregate
5 Inpatient morbidity and mortality eBAyeGv0exc aggregate
6 Malaria case diagnosis, treatment and investigation qDkgAbB5Jlk tracker
7 Malaria case registration VBqh0ynB2wv aggregate
8 Malaria focus investigation M3xtLkYBlKI tracker
9 Malaria testing and surveillance bMcwwoVnbSR aggregate
10 MNCH / PNC (Adult Woman) uy2gU8kT1jF tracker
11 Provider Follow-up and Support Tool fDd25txQckK tracker
12 TB program ur1Edk5Oe2n tracker
13 WHO RMNCH Tracker WSGAb5XwJ3Y tracker
14 XX MAL RDT - Case Registration MoUd5BTQ3lY aggregateR
# get the names and IDs of the organisation units
demo_units <- readepi::get_organisation_units(login = demo_login)
tibble::as_tibble(demo_units)
OUTPUT
# A tibble: 1,166 × 8
National_name National_id District_name District_id Chiefdom_name Chiefdom_id
<chr> <chr> <chr> <chr> <chr> <chr>
1 Sierra Leone ImspTQPwCqd Western Area at6UHUQatSo Rural Wester… qtr8GGlm4gg
2 Sierra Leone ImspTQPwCqd Western Area at6UHUQatSo Rural Wester… qtr8GGlm4gg
3 Sierra Leone ImspTQPwCqd Bo O6uvpzGd5pu Kakua U6Kr7Gtpidn
4 Sierra Leone ImspTQPwCqd Kambia PMa2VCrupOd Magbema QywkxFudXrC
5 Sierra Leone ImspTQPwCqd Tonkolili eIQbndfxQMb Yoni NNE0YMCDZkO
6 Sierra Leone ImspTQPwCqd Port Loko TEQlaapDQoK Kaffu Bullom vn9KJsLyP5f
7 Sierra Leone ImspTQPwCqd Koinadugu qhqAxPSTUXp Nieni J4GiUImJZoE
8 Sierra Leone ImspTQPwCqd Western Area at6UHUQatSo Freetown C9uduqDZr9d
9 Sierra Leone ImspTQPwCqd Western Area at6UHUQatSo Freetown C9uduqDZr9d
10 Sierra Leone ImspTQPwCqd Kono Vth0fbpFcsO Gbense TQkG0sX9nca
# ℹ 1,156 more rows
# ℹ 2 more variables: Facility_name <chr>, Facility_id <chr>Importing data from SORMAS
The SORMAS (Surveillance Outbreak
Response Management and Analysis System) is an open-source e-health
system that optimizes infectious disease surveillance and outbreak
response processes. The readepi::read_sormas() function
allows you to import data from SORMAS via its API.
In the current version of the readepi package, the
read_sormas() function returns data for the following
columns: case_id, person_id, sex, date_of_birth, case_origin,
country, city, lat, long, case_status, date_onset, date_admission,
date_last_contact, date_first_contact, outcome, date_outcome,
and Ct_values.
The code chunk below demonstrates how to import data from a demo SORMAS system:
R
# CONNECT TO THE SORMAS SYSTEM
sormas_login <- readepi::login(
type = "sormas",
from = "https://demo.sormas.org/sormas-rest",
user_name = "SurvSup",
password = "Lk5R7JXeZSEc"
)
# FETCH ALL COVID (Corona virus) CASES FROM THE TEST SORMAS INSTANCE
covid_cases <- readepi::read_sormas(
login = sormas_login,
disease = "coronavirus",
)
tibble::as_tibble(covid_cases)
OUTPUT
# A tibble: 2 × 15
case_id person_id date_onset case_origin case_status outcome sex
<chr> <chr> <date> <chr> <chr> <chr> <chr>
1 WJHHRV-A2UPKG-R37W… RYBAQX-A… 2025-11-11 IN_COUNTRY NOT_CLASSI… NO_OUT… <NA>
2 VPMCMM-YUZENC-P3JN… U2BJQK-M… 2025-11-01 IN_COUNTRY SUSPECT DECEAS… <NA>
# ℹ 8 more variables: date_of_birth <chr>, country <chr>, city <chr>,
# latitude <chr>, longitude <chr>, contact_id <chr>,
# date_last_contact <date>, Ct_values <chr>A key parameter is the disease name. To ensure correct syntax, you
can retrieve the list of available disease names using the
sormas_get_diseases() function.
R
# get the list of all disease names
disease_names <- readepi::sormas_get_diseases(
login = sormas_login
)
tibble::as_tibble(disease_names)
OUTPUT
# A tibble: 67 × 2
disease active
<chr> <chr>
1 AFP TRUE
2 CHOLERA TRUE
3 CONGENITAL_RUBELLA TRUE
4 DENGUE TRUE
5 EVD TRUE
6 GUINEA_WORM TRUE
7 LASSA TRUE
8 MEASLES TRUE
9 MONKEYPOX TRUE
10 NEW_INFLUENZA TRUE
# ℹ 57 more rowsReading from Demo SORMAS sever
The SORMAS organization also provides demo servers for development and testing. One of these is called clinical surveillance, available at the link (“https://demo.sormas.org/sormas-rest”), and accessible with username “CaseSup” and password “SJgFKffPDmr7”. Log into this server, list all available diseases, and import cases related to the monkeypox (mpox) disease.
R
# establish the connection to the system
sormas_demo <- readepi::login(
type = "sormas",
from = "https://demo.sormas.org/sormas-rest",
user_name = "CaseSup",
password = "SJgFKffPDmr7"
)
# List the names of all disease
demo_diseases <- readepi::sormas_get_diseases(login = sormas_demo)
tibble::as_tibble(demo_diseases)
OUTPUT
# A tibble: 67 × 2
disease active
<chr> <chr>
1 AFP TRUE
2 CHOLERA TRUE
3 CONGENITAL_RUBELLA TRUE
4 DENGUE TRUE
5 EVD TRUE
6 GUINEA_WORM TRUE
7 LASSA TRUE
8 MEASLES TRUE
9 MONKEYPOX TRUE
10 NEW_INFLUENZA TRUE
# ℹ 57 more rowsR
# get the list of all disease names
mpox_cases <- readepi::read_sormas(
login = sormas_demo,
disease = "monkeypox",
)
WARNING
Warning in as.POSIXct(as.numeric(x), origin = "1970-01-01"): NAs introduced by
coercionR
tibble::as_tibble(mpox_cases)
OUTPUT
# A tibble: 1 × 15
case_id person_id date_onset case_origin case_status outcome sex
<chr> <chr> <date> <chr> <chr> <chr> <chr>
1 WQLS6O-ZEZ562-MAGM… WVADAF-S… NA IN_COUNTRY PROBABLE NO_OUT… <NA>
# ℹ 8 more variables: date_of_birth <chr>, country <chr>, city <chr>,
# latitude <chr>, longitude <chr>, contact_id <chr>,
# date_last_contact <date>, Ct_values <chr>Key Points
- Use rio, io, readr or
{ImportExport}to read data from individual files. - Use readepi to read data from RDBMS and HIS.
- The {rio} package supports a wide range of file formats including
CSV,TSV,XLSX, and compressed files. - Use
readepi::login()to establish connections to RDBMS, DHIS2, or SORMAS systems. - The readepi package currently supports importing data from DHIS2 and SORMAS health information systems.
Content from Clean case data
Last updated on 2026-02-24 | Edit this page
Overview
Questions
- How to clean and standardize case data?
Objectives
- Explain how to clean, curate, and standardize case data using cleanepi package
- Perform essential data-cleaning operations on a real case dataset.
Prerequisite
In this episode, we will use a simulated Ebola dataset that can be:
- Download the simulated_ebola_2.csv
- Save it in the
data/folder. Follow instructions in Setup to configure an RStudio Project and folder
Introduction
In the process of analyzing outbreak data, it’s essential to ensure that the dataset is clean, curated, standardized, and validated. This will ensure that analysis is accurate (i.e. you are analysing what you think you are analysing) and reproducible (i.e. if someone wants to go back and repeat your analysis steps with your code, you can be confident they will get the same results). This episode focuses on cleaning epidemics and outbreaks data using the cleanepi package, For demonstration purposes, we’ll work with a simulated dataset of Ebola cases.
Let’s start by loading the package rio to read data
and the package cleanepi to clean it. We’ll use the pipe
%>% to connect some of their functions, including others
from the package dplyr, so let’s also call to the
tidyverse package:
R
# Load packages
library(tidyverse) # for {dplyr} functions and the pipe %>%
library(rio) # for importing data
library(here) # for easy file referencing
library(cleanepi)
The double-colon (::)
operator
The :: in R lets you access functions or objects from a
specific package without attaching the entire package to the search
path. It offers several important advantages including the
followings:
- Telling explicitly which package a function comes from, reducing ambiguity and potential conflicts when several packages have functions with the same name.
- Allowing to call a function from a package without loading the whole package with library().
For example, the command dplyr::filter(data, condition)
means we are calling the filter() function from the
dplyr package.
The first step is to import the dataset into working environment.
This can be done by following the guidelines outlined in the Read case data episode. It involves loading
the dataset into R environment and view its structure and
content.
R
# Read data
# e.g.: if path to file is data/simulated_ebola_2.csv then:
raw_ebola_data <- rio::import(
here::here("data", "simulated_ebola_2.csv")
) %>%
dplyr::as_tibble() # for a simple data frame output
R
# Print data frame
raw_ebola_data
OUTPUT
# A tibble: 15,003 × 9
V1 `case id` age gender status `date onset` `date sample` lab region
<int> <int> <chr> <chr> <chr> <chr> <chr> <lgl> <chr>
1 1 14905 90 1 "conf… 03/15/2015 06/04/2015 NA valdr…
2 2 13043 twenty… 2 "" Sep /11/13 03/01/2014 NA valdr…
3 3 14364 54 f <NA> 09/02/2014 03/03/2015 NA valdr…
4 4 14675 ninety <NA> "" 10/19/2014 31/ 12 /14 NA valdr…
5 5 12648 74 F "" 08/06/2014 10/10/2016 NA valdr…
6 5 12648 74 F "" 08/06/2014 10/10/2016 NA valdr…
7 6 14274 sevent… female "" Apr /05/15 01/23/2016 NA valdr…
8 7 14132 sixteen male "conf… Dec /29/Y 05/10/2015 NA valdr…
9 8 14715 44 f "conf… Apr /06/Y 04/24/2016 NA valdr…
10 9 13435 26 1 "" 09/07/2014 20/ 09 /14 NA valdr…
# ℹ 14,993 more rowsDiscussion
Let’s first diagnose the data frame. List all the characteristics in the data frame above that are problematic for data analysis.
Are any of those characteristics familiar from any previous data analysis you have performed?
A quick inspection
Quick exploration and inspection of the dataset are crucial to
identify potential data issues before diving into any analysis tasks.
The cleanepi package simplifies this process with the
scan_data() function. Let’s take a look at how you can use
it:
R
cleanepi::scan_data(raw_ebola_data)
OUTPUT
Field_names missing numeric date character logical
1 age 0.0690 0.8925 0.0000 0.1075 0
2 gender 0.1874 0.0560 0.0000 0.9440 0
3 status 0.0565 0.0000 0.0000 1.0000 0
4 date onset 0.0001 0.0000 0.9159 0.0841 0
5 date sample 0.0001 0.0000 1.0000 0.0000 0
6 region 0.0000 0.0000 0.0000 1.0000 0The results provide an overview of the content of all character columns, including column names, and the percent of some data types within them. You can see that the column names in the dataset are descriptive but lack consistency. Some are composed of multiple words separated by white spaces. Additionally, some columns contain more than one data type, and there are missing values in the form of an empty string in others.
Common operations
This section demonstrate how to perform some common data cleaning operations using the cleanepi package.
Standardizing column names
For this example dataset, standardizing column names typically
involves removing with spaces and connecting different words with “_”.
This practice helps maintain consistency and readability in the dataset.
However, the function used for standardizing column names offers more
options. Type ?cleanepi::standardize_column_names in the
console for more details.
R
sim_ebola_data <- cleanepi::standardize_column_names(raw_ebola_data)
names(sim_ebola_data)
OUTPUT
[1] "v1" "case_id" "age" "gender" "status"
[6] "date_onset" "date_sample" "lab" "region" If you want to maintain certain column names without subjecting them
to the standardization process, you can utilize the keep
argument of the function
cleanepi::standardize_column_names(). This argument accepts
a vector of column names that are intended to be kept unchanged.
Challenge
What differences can you observe in the column names?
Standardize the column names of the input dataset, but keep the first column names as it is.
You can try
cleanepi::standardize_column_names(data = raw_ebola_data, keep = "V1")
Removing irregularities
Raw data may contain fields that don’t add any variability to the
data such as empty rows and columns, or
constant columns (where all entries have the same
value). It can also contain duplicated rows. Functions
from cleanepi like remove_duplicates() and
remove_constants() remove such irregularities as
demonstrated in the below code chunk.
R
# Remove constants
sim_ebola_data <- cleanepi::remove_constants(sim_ebola_data)
Now, print the output to identify what constant column you removed!
R
# Remove duplicates
sim_ebola_data <- cleanepi::remove_duplicates(sim_ebola_data)
OUTPUT
! Found 5 duplicated rows in the dataset.
ℹ Use `print_report(dat, "found_duplicates")` to access them, where "dat" is
the object used to store the output from this operation.You can get the number and location of the duplicated rows that where
found. Run cleanepi::print_report(), wait for the report to
open in your browser, and find the “Duplicates” tab.
To use this information within R, you can print data frames with
specific sections of the report in the console using the argument
what.
R
# Print a report of found duplicates
cleanepi::print_report(data = sim_ebola_data, what = "found_duplicates")
# Print a report of removed duplicates
cleanepi::print_report(data = sim_ebola_data, what = "removed_duplicates")
Challenge
In the following data frame:
OUTPUT
# A tibble: 6 × 5
col1 col2 col3 col4 col5
<dbl> <dbl> <chr> <chr> <date>
1 1 1 a b NA
2 2 3 a b NA
3 NA NA a <NA> NA
4 NA NA a <NA> NA
5 NA NA a <NA> NA
6 NA NA <NA> <NA> NA What columns are the:
- constant data?
- duplicated rows?
Constant data mostly refers to empty rows or columns as well as constant columns.
Replacing missing values
In addition to the irregularities, raw data may contain missing
values, and these may be encoded by different strings
(e.g. "NA", "", character(0)). To
ensure robust analysis, it is a good practice to replace all missing
values by NA in the entire dataset. Below is a code snippet
demonstrating how you can achieve this in cleanepi for
missing entries represented by an empty string "":
R
sim_ebola_data <- cleanepi::replace_missing_values(
data = sim_ebola_data,
na_strings = ""
)
sim_ebola_data
OUTPUT
# A tibble: 15,000 × 7
v1 case_id age gender status date_onset date_sample
<int> <int> <chr> <chr> <chr> <chr> <chr>
1 1 14905 90 1 confirmed 03/15/2015 06/04/2015
2 2 13043 twenty-five 2 <NA> sep /11/13 03/01/2014
3 3 14364 54 f <NA> 09/02/2014 03/03/2015
4 4 14675 ninety <NA> <NA> 10/19/2014 31/ 12 /14
5 5 12648 74 F <NA> 08/06/2014 10/10/2016
6 6 14274 seventy-six female <NA> apr /05/15 01/23/2016
7 7 14132 sixteen male confirmed dec /29/y 05/10/2015
8 8 14715 44 f confirmed apr /06/y 04/24/2016
9 9 13435 26 1 <NA> 09/07/2014 20/ 09 /14
10 10 14816 thirty f <NA> 06/29/2015 06/02/2015
# ℹ 14,990 more rowsValidating subject IDs
Each entry in the dataset represents a subject (e.g. a disease case
or study participant) and should be distinguishable by a specific ID
formatted in a particular way. These IDs can contain numbers falling
within a specific range, a prefix and/or suffix, and might be written
such that they contain a specific number of characters. The
cleanepi package offers the function
check_subject_ids() designed precisely for this task as
shown in the below code chunk. This function checks whether the IDs are
unique and meet the required criteria specified by the user.
R
# check if the subject IDs in the 'case_id' column contains numbers ranging
# from 0 to 15000
sim_ebola_data <- cleanepi::check_subject_ids(
data = sim_ebola_data,
target_columns = "case_id",
range = c(0, 15000)
)
OUTPUT
! Detected 0 missing, 1957 duplicated, and 0 incorrect subject IDs.
ℹ Enter `print_report(data = dat, "incorrect_subject_id")` to access them,
where "dat" is the object used to store the output from this operation.
ℹ You can use the `correct_subject_ids()` function to correct them.Note that our simulated dataset contains duplicated subject IDs.
Let’s print a preliminary report with
cleanepi::print_report(sim_ebola_data). Focus on the
“Unexpected subject ids” tab to identify what IDs require an extra
treatment.
In the console, you can print:
R
print_report(data = sim_ebola_data, "incorrect_subject_id")
After finishing this tutorial, we invite you to explore the package reference guide of cleanepi::check_subject_ids() to find the function that can fix this situation.
Standardizing dates
An epidemic dataset typically contains date columns for different events, such as the date of infection, date of symptoms onset, etc. These dates can come in different date formats, and it is good practice to standardize them to benefit from the powerful R functionalities designed to handle date values in downstream analyses. The cleanepi package provides functionality for converting date columns of epidemic datasets into ISO8601 format, ensuring consistency across the different date columns. Here’s how you can use it on our simulated dataset:
R
sim_ebola_data <- cleanepi::standardize_dates(
sim_ebola_data,
target_columns = c("date_onset", "date_sample")
)
OUTPUT
! Detected 1142 values that comply with multiple formats and no values that are
outside of the specified time frame.
ℹ Enter `print_report(data = dat, "date_standardization")` to access them,
where "dat" is the object used to store the output from this operation.R
sim_ebola_data
OUTPUT
# A tibble: 15,000 × 7
v1 case_id age gender status date_onset date_sample
<int> <chr> <chr> <chr> <chr> <date> <date>
1 1 14905 90 1 confirmed 2015-03-15 2015-04-06
2 2 13043 twenty-five 2 <NA> 2013-09-11 2014-01-03
3 3 14364 54 f <NA> 2014-02-09 2015-03-03
4 4 14675 ninety <NA> <NA> 2014-10-19 2014-12-31
5 5 12648 74 F <NA> 2014-06-08 2016-10-10
6 6 14274 seventy-six female <NA> 2015-04-05 2016-01-23
7 7 14132 sixteen male confirmed NA 2015-10-05
8 8 14715 44 f confirmed NA 2016-04-24
9 9 13435 26 1 <NA> 2014-07-09 2014-09-20
10 10 14816 thirty f <NA> 2015-06-29 2015-02-06
# ℹ 14,990 more rowsThis function converts the values in the target columns into the YYYY-mm-dd format.
How is this possible?
We invite you to find the key package that makes this standardisation possible inside cleanepi by reading the “Details” section of the Standardize date variables reference manual!
Also, check how to use the orders argument if you want
to target US format character strings. You can explore this
reproducible example.
Converting to numeric values
In the raw dataset, some columns can come with mixture of character
and numerical values, and you will often want to convert character
values for numbers explicitly into numeric values
(e.g. "seven" to 7). For example, in our
simulated data set, in the age column some entries are written in words.
In cleanepi the function
convert_to_numeric() does such conversion as illustrated in
the below code chunk.
R
sim_ebola_data <- cleanepi::convert_to_numeric(
data = sim_ebola_data,
target_columns = "age"
)
sim_ebola_data
OUTPUT
# A tibble: 15,000 × 7
v1 case_id age gender status date_onset date_sample
<int> <chr> <dbl> <chr> <chr> <date> <date>
1 1 14905 90 1 confirmed 2015-03-15 2015-04-06
2 2 13043 25 2 <NA> 2013-09-11 2014-01-03
3 3 14364 54 f <NA> 2014-02-09 2015-03-03
4 4 14675 90 <NA> <NA> 2014-10-19 2014-12-31
5 5 12648 74 F <NA> 2014-06-08 2016-10-10
6 6 14274 76 female <NA> 2015-04-05 2016-01-23
7 7 14132 16 male confirmed NA 2015-10-05
8 8 14715 44 f confirmed NA 2016-04-24
9 9 13435 26 1 <NA> 2014-07-09 2014-09-20
10 10 14816 30 f <NA> 2015-06-29 2015-02-06
# ℹ 14,990 more rowsMultiple language support
Thanks to the numberize package, we can convert numbers written in English, French or Spanish into positive integer values.
Epidemiology related operations
In addition to common data cleansing tasks, such as those discussed in the above section, the cleanepi package offers additional functionalities tailored specifically for processing and analyzing outbreak and epidemic data. This section covers some of these specialized tasks.
Checking sequence of dated-events
Ensuring the correct order and sequence of dated events is crucial in
epidemiological data analysis, especially when analyzing infectious
diseases where the timing of events like symptom onset and sample
collection is essential. The cleanepi package provides a
helpful function called check_date_sequence() designed for
this purpose.
Here’s an example of a code chunk demonstrating the usage of the
function check_date_sequence() in the first 100 records of
our simulated Ebola dataset.
R
cleanepi::check_date_sequence(
data = sim_ebola_data[1:100, ],
target_columns = c("date_onset", "date_sample")
)
OUTPUT
ℹ Cannot check the sequence of date events across 37 rows due to missing data.OUTPUT
! Detected 24 incorrect date sequences at lines: "8, 15, 18, 20, 21, 23, 26,
28, 29, 32, 34, 35, 37, 38, 40, 43, 46, 49, 52, 54, 56, 58, 60, 63".
ℹ Enter `print_report(data = dat, "incorrect_date_sequence")` to access them,
where "dat" is the object used to store the output from this operation.This functionality is crucial for ensuring data integrity and accuracy in epidemiological analyses, as it helps identify any inconsistencies or errors in the chronological order of events, allowing you to address them appropriately.
Dictionary-based substitution
In the realm of data pre-processing, it’s common to encounter scenarios where certain columns in a dataset, such as the “gender” column in our simulated Ebola dataset, are expected to have specific values or factors. However, it’s also common for unexpected or erroneous values to appear in these columns, which need to be replaced with the appropriate values. The cleanepi package offers support for dictionary-based substitution, a method that allows you to replace values in specific columns based on mappings defined in a data dictionary. This approach ensures consistency and accuracy in data cleaning.
Moreover, cleanepi provides a built-in dictionary specifically tailored for epidemiological data. The example dictionary below includes mappings for the “gender” column.
R
test_dict <- base::readRDS(
system.file("extdata", "test_dict.RDS", package = "cleanepi")
) %>%
dplyr::as_tibble()
test_dict
OUTPUT
# A tibble: 6 × 4
options values grp orders
<chr> <chr> <chr> <int>
1 1 male gender 1
2 2 female gender 2
3 M male gender 3
4 F female gender 4
5 m male gender 5
6 f female gender 6Now, we can use this dictionary to standardize values of the the
“gender” column according to predefined categories. Below is an example
code chunk demonstrating how to perform this using the
clean_using_dictionary() function from the {cleanepi}
package.
R
sim_ebola_data <- cleanepi::clean_using_dictionary(
data = sim_ebola_data,
dictionary = test_dict
)
sim_ebola_data
OUTPUT
# A tibble: 15,000 × 7
v1 case_id age gender status date_onset date_sample
<int> <chr> <dbl> <chr> <chr> <date> <date>
1 1 14905 90 male confirmed 2015-03-15 2015-04-06
2 2 13043 25 female <NA> 2013-09-11 2014-01-03
3 3 14364 54 female <NA> 2014-02-09 2015-03-03
4 4 14675 90 <NA> <NA> 2014-10-19 2014-12-31
5 5 12648 74 female <NA> 2014-06-08 2016-10-10
6 6 14274 76 female <NA> 2015-04-05 2016-01-23
7 7 14132 16 male confirmed NA 2015-10-05
8 8 14715 44 female confirmed NA 2016-04-24
9 9 13435 26 male <NA> 2014-07-09 2014-09-20
10 10 14816 30 female <NA> 2015-06-29 2015-02-06
# ℹ 14,990 more rowsThis approach simplifies the data cleaning process, ensuring that categorical variables in epidemiological datasets are accurately categorized and ready for further analysis.
Note that, when a column in the dataset contains values that are not
in the dictionary, the function
cleanepi::clean_using_dictionary() will raise an error. You
can start a custom dictionary with a data frame inside or outside R and
use the function cleanepi::add_to_dictionary() to include
new elements in the dictionary. For example:
R
new_dictionary <- tibble::tibble(
options = "0",
values = "female",
grp = "sex",
orders = 1L
) %>%
cleanepi::add_to_dictionary(
option = "1",
value = "male",
grp = "sex",
order = NULL
)
new_dictionary
OUTPUT
# A tibble: 2 × 4
options values grp orders
<chr> <chr> <chr> <int>
1 0 female sex 1
2 1 male sex 2You can have more details in the section about “Dictionary-based data substituting” in the package vignette.
Calculating time span between different date events
In epidemiological data analysis, it is also useful to track and analyze time-dependent events, such as the progression of a disease outbreak (i.e., the time elapsed between today and the date the first case was reported) or the duration between date of sample collection and analysis (i.e., the time difference between today and the sample collection date). The most common example is to calculate the age of all the subjects given their dates of birth (i.e., the time difference between today and their date of birth).
The cleanepi package offers a convenient function for
calculating the time elapsed between two dated events at different time
scales. For example, the below code snippet utilizes the function
cleanepi::timespan() to compute the time elapsed since the
date of sampling of the identified cases until the \(3^{rd}\) of January 2025
("2025-01-03").
R
sim_ebola_data <- cleanepi::timespan(
data = sim_ebola_data,
target_column = "date_sample",
end_date = as.Date("2025-01-03"),
span_unit = "years",
span_column_name = "years_since_collection",
span_remainder_unit = "months"
)
sim_ebola_data %>%
dplyr::select(case_id, date_sample, years_since_collection, remainder_months)
OUTPUT
# A tibble: 15,000 × 4
case_id date_sample years_since_collection remainder_months
<chr> <date> <dbl> <dbl>
1 14905 2015-04-06 9 8
2 13043 2014-01-03 11 0
3 14364 2015-03-03 9 10
4 14675 2014-12-31 10 0
5 12648 2016-10-10 8 2
6 14274 2016-01-23 8 11
7 14132 2015-10-05 9 2
8 14715 2016-04-24 8 8
9 13435 2014-09-20 10 3
10 14816 2015-02-06 9 10
# ℹ 14,990 more rowsAfter executing the function cleanepi::timespan(), two
new columns named years_since_collection and
remainder_months are added to the
sim_ebola_data dataset. For each case, these columns
respectively represent the calculated time elapsed since the date of
sample collection measured in years, and the remaining time measured in
months.
Challenge
Age data is useful in many downstream analysis. You can categorize it to generate stratified estimates.
Read the test_df.RDS data frame within the
cleanepi package:
R
dat <- readRDS(
file = system.file("extdata", "test_df.RDS", package = "cleanepi")
) %>%
dplyr::as_tibble()
Calculate the age in years until today’s date of the subjects from their date of birth, and the remainder time in months. Clean and standardize the required elements to get this done.
Before calculating the age, you may need to:
- standardize column names
- standardize dates columns
- replace missing value strings with NA
In the solution we added date_first_pcr_positive_test as
part of the Date columns to be standardised given that it often used in
disease outbreak analysis.
R
dat_clean <- dat %>%
# standardize column names and dates
cleanepi::standardize_column_names() %>%
cleanepi::standardize_dates(
target_columns = c("date_of_birth", "date_first_pcr_positive_test")
) %>%
# replace missing value strings with NA
cleanepi::replace_missing_values(
target_columns = c("sex", "date_of_birth"),
na_strings = "-99"
) %>%
# calculate the age in 'years' and return the remainder in 'months'
cleanepi::timespan(
target_column = "date_of_birth",
end_date = Sys.Date(),
span_unit = "years",
span_column_name = "age_in_years",
span_remainder_unit = "months"
)
OUTPUT
! Detected 4 values that comply with multiple formats and no values that are
outside of the specified time frame.
ℹ Enter `print_report(data = dat, "date_standardization")` to access them,
where "dat" is the object used to store the output from this operation.
! Found <numeric> values that could also be of type <Date> in column:
date_of_birth.
ℹ It is possible to convert them into <Date> using: `lubridate::as_date(x,
origin = as.Date("1900-01-01"))`
• where "x" represents here the vector of values from these columns
(`data$target_column`).Now, How would you categorize a numerical variable?
The simplest alternative is using Hmisc::cut2(). You can
also use dplyr::case_when(). However, this requires more
lines of code and is more appropriate for custom categorization. Here we
provide one solution using base::cut():
R
dat_clean <- dat_clean %>%
# select the columns of interest
dplyr::select(
study_id,
sex,
date_first_pcr_positive_test,
date_of_birth,
age_in_years
) %>%
# categorize the age variable [add as a challenge hint]
dplyr::mutate(
age_category = base::cut(
x = age_in_years,
breaks = c(0, 20, 35, 60, Inf), # replace with max value if known
include.lowest = TRUE,
right = FALSE
)
)
You can investigate the maximum values of variables from the summary
made from the skimr::skim() function. Instead of
base::cut() you can also use
Hmisc::cut2(x = age_in_years, cuts = c(20,35,60)), which
gives the maximum value and do not require more arguments.
Multiple operations at once
Performing data cleaning operations individually can be
time-consuming and error-prone. The cleanepi package
simplifies this process by offering a convenient wrapper function called
clean_data(), which allows you to perform multiple
operations at once.
When no cleaning operation is specified, the
clean_data() function automatically applies a series of
data cleaning operations to the input dataset. Here’s an example code
chunk illustrating how to use clean_data() on a raw
simulated Ebola dataset:
R
cleaned_data <- cleanepi::clean_data(raw_ebola_data)
OUTPUT
ℹ Cleaning column namesOUTPUT
ℹ Removing constant columns and empty rowsOUTPUT
ℹ Removing duplicated rowsOUTPUT
! Found 5 duplicated rows in the dataset.
ℹ Use `print_report(dat, "found_duplicates")` to access them, where "dat" is
the object used to store the output from this operation.Further more, you can combine multiple data cleaning tasks via the
base R pipe (%>%) or the {magrittr} pipe
(%>%) operator, as shown in the below code snippet.
R
# Perform the cleaning operations using the pipe (%>%) operator
cleaned_data <- raw_ebola_data %>%
cleanepi::standardize_column_names() %>%
cleanepi::remove_constants() %>%
cleanepi::remove_duplicates() %>%
cleanepi::replace_missing_values(na_strings = "") %>%
cleanepi::check_subject_ids(
target_columns = "case_id",
range = c(1, 15000)
) %>%
cleanepi::standardize_dates(
target_columns = c("date_onset", "date_sample")
) %>%
cleanepi::convert_to_numeric(target_columns = "age") %>%
cleanepi::check_date_sequence(
target_columns = c("date_onset", "date_sample")
) %>%
cleanepi::clean_using_dictionary(dictionary = test_dict) %>%
cleanepi::timespan(
target_column = "date_sample",
end_date = as.Date("2025-01-03"),
span_unit = "years",
span_column_name = "years_since_collection",
span_remainder_unit = "months"
)
Challenge
Have you noticed that cleanepi contains a set of functions to diagnose the cleaning status of the dataset and another set to perform cleaning actions on it?
To identify both groups:
- On a piece of paper, write the names of each function under the corresponding column:
| Diagnose cleaning status | Perform cleaning action |
|---|---|
| … | … |
Cleaning report
The cleanepi package generates a comprehensive report detailing the findings and actions of all data cleansing operations conducted during the analysis. This report is presented as a HTML file that automatically opens in your browser with. Each section corresponds to a specific data cleansing operation, and clicking on each section allows you to access the results of that particular operation. This interactive approach enables users to efficiently review and analyze the effects of individual cleansing steps within the broader data cleansing process.
You can view the report using:
R
cleanepi::print_report(data = cleaned_data)
Example of data cleaning report generated by cleanepi
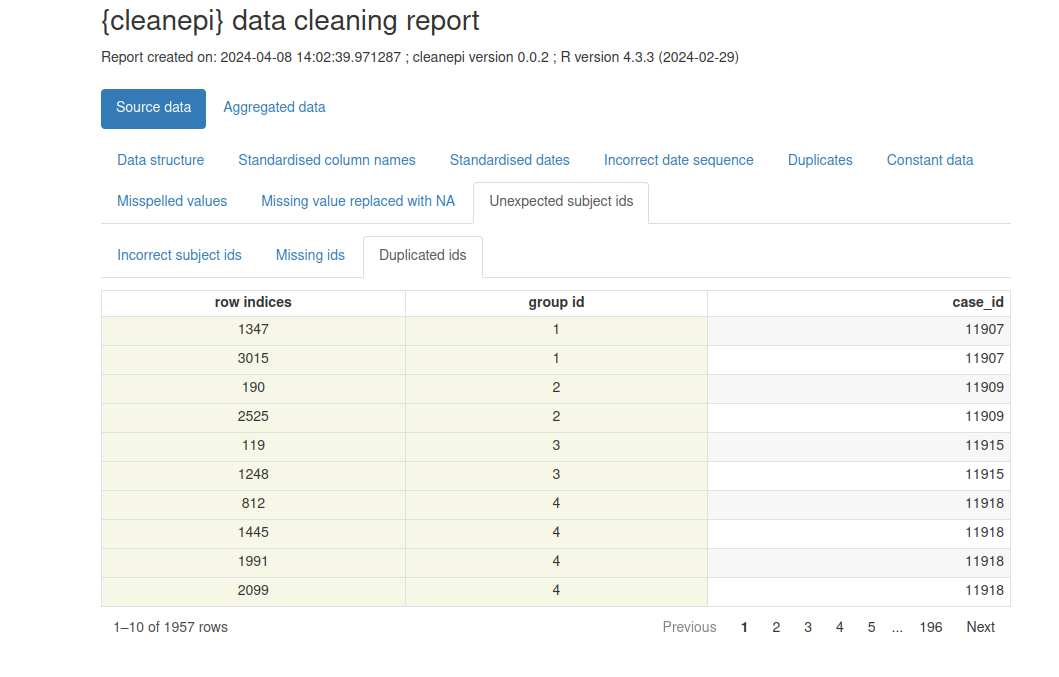
Content from Validate case data
Last updated on 2026-02-24 | Edit this page
Overview
Questions
- How to convert a raw dataset into a
linelistobject?
Objectives
- Demonstrate how to covert case data into
linelistdata - Demonstrate how to tag and validate data to make analysis more reliable
Prerequisite
This episode requires you to:
- Download the cleaned_data.csv file
- and save it in the
data/folder.
Introduction
In outbreak analysis, once you have completed the initial steps of
reading and cleaning the case data, it’s essential to establish an
additional fundamental layer to ensure the integrity and reliability of
subsequent analyses. Otherwise you might encounter issues during the
analysis process due to creation or removal of specific variables,
changes in their underlying data types (like <date>
or <chr>), etc. Specifically, this additional step
involves:
- Verifying the presence and correct data type of certain columns within your dataset, a process commonly referred to as tagging;
- Implementing measures to make sure that these tagged columns are not inadvertently deleted during further data processing steps, known as validation.
This episode focuses on tagging and validating outbreak data using
the linelist
package. Let’s start by loading the package rio to read
data and the linelist package to create a linelist
object. We’ll use the pipe operator (%>%) to connect
some of their functions, including others from the package
dplyr. For this reason, we will also load the {tidyverse}
package.
R
# Load packages
library(tidyverse) # to access {dplyr} functions and the pipe %>% operator
# from {magrittr}
library(rio) # for importing data
library(here) # for easy file referencing
library(linelist) # for tagging and validating
The double-colon (::)
operator
The::in R lets you access functions or objects from a
specific package without attaching the entire package to the search
path. It offers several important advantages including the
followings:
- Telling explicitly which package a function comes from, reducing ambiguity and potential conflicts when several packages have functions with the same name.
- Allowing to call a function from a package without loading the whole package with library().
For example, the command dplyr::filter(data, condition)
means we are calling the filter() function from the
dplyr package.
Import the dataset following the guidelines outlined in the Read case data episode. This involves loading the dataset into the working environment and view its structure and content.
R
# Read data
# e.g.: if path to file is data/simulated_ebola_2.csv then:
cleaned_data <- rio::import(
here::here("data", "cleaned_data.csv")
) %>%
dplyr::as_tibble() # for a simple data frame output
OUTPUT
# A tibble: 15,000 × 9
v1 case_id age gender status date_onset date_sample
<int> <int> <dbl> <chr> <chr> <IDate> <IDate>
1 1 14905 90 male confirmed 2015-03-15 2015-04-06
2 2 13043 25 female <NA> 2013-09-11 2014-01-03
3 3 14364 54 female <NA> 2014-02-09 2015-03-03
4 4 14675 90 <NA> <NA> 2014-10-19 2014-12-31
5 5 12648 74 female <NA> 2014-06-08 2016-10-10
6 6 14274 76 female <NA> 2015-04-05 2016-01-23
7 7 14132 16 male confirmed NA 2015-10-05
8 8 14715 44 female confirmed NA 2016-04-24
9 9 13435 26 male <NA> 2014-07-09 2014-09-20
10 10 14816 30 female <NA> 2015-06-29 2015-02-06
# ℹ 14,990 more rows
# ℹ 2 more variables: years_since_collection <int>, remainder_months <int>Discussion
An unexpected change
You are in an emergency response situation. You need to generate daily situation reports. You automated your analysis to read data directly from the online server 😁. However, the people in charge of the data collection/administration needed to remove/rename/reformat one variable you found helpful 😞!
How can you detect if the input data is still valid to replicate the analysis code you wrote the day before?
Creating a linelist and tagging columns
Once the data is loaded and cleaned, we can convert the cleaned case
data into a linelist object using linelist
package, as in the below code chunk.
R
# Create a linelist object from cleaned data
linelist_data <- linelist::make_linelist(
x = cleaned_data, # Input data
id = "case_id", # Column for unique case identifiers
date_onset = "date_onset", # Column for date of symptom onset
gender = "gender" # Column for gender
)
# Display the resulting linelist object
linelist_data
OUTPUT
// linelist object
# A tibble: 15,000 × 9
v1 case_id age gender status date_onset date_sample
<int> <int> <dbl> <chr> <chr> <IDate> <IDate>
1 1 14905 90 male confirmed 2015-03-15 2015-04-06
2 2 13043 25 female <NA> 2013-09-11 2014-01-03
3 3 14364 54 female <NA> 2014-02-09 2015-03-03
4 4 14675 90 <NA> <NA> 2014-10-19 2014-12-31
5 5 12648 74 female <NA> 2014-06-08 2016-10-10
6 6 14274 76 female <NA> 2015-04-05 2016-01-23
7 7 14132 16 male confirmed NA 2015-10-05
8 8 14715 44 female confirmed NA 2016-04-24
9 9 13435 26 male <NA> 2014-07-09 2014-09-20
10 10 14816 30 female <NA> 2015-06-29 2015-02-06
# ℹ 14,990 more rows
# ℹ 2 more variables: years_since_collection <int>, remainder_months <int>
// tags: id:case_id, date_onset:date_onset, gender:gender The linelist package supplies tags for common
epidemiological variables and a set of appropriate data types for each.
You can view the list of available tags by the variable name and their
acceptable data types using the linelist::tags_types()
function.
Challenge
Let’s tag more variables. In some datasets, it is possible to encounter variable names that are different from the available tag names. In such cases, we can associate them based on how variables were defined for data collection.
Now: -Explore the available tag names in
linelist. -Find what other variables in
the input dataset can be associated with any of these available tags.
-Tag those variables as shown above using the
linelist::make_linelist() function.
Your can get access to the list of available tag names in linelist using:
R
# Get a list of available tags names and data types
linelist::tags_types()
# Get a list of names only
linelist::tags_names()
R
linelist::make_linelist(
x = cleaned_data,
id = "case_id",
date_onset = "date_onset",
gender = "gender",
age = "age",
# same name in default list and dataset
date_reporting = "date_sample" # different names but related
)
Are these additional tags visible in the output?
< !–Do you want to see a display of available and tagged
variables? You can explore the function linelist::tags()
and read its reference
documentation.- ->
Validation
To ensure that all tagged variables are standardized and have the
correct data types, use the linelist::validate_linelist()
function, as shown in the example below:
R
linelist::validate_linelist(linelist_data)
OUTPUT
'linelist_data' is a valid linelist objectIf your dataset requires a new tag other than those defined in the
linelist package, use allow_extra = TRUE
when creating the linelist object with its corresponding datatype using
the linelist::make_linelist() function.
Challenge
Let’s assume the following scenario during an ongoing outbreak. You notice at some point that the data stream you have been relying on has a set of new entries (i.e., rows or observations), and the data type of one variable has changed.
Let’s consider the example where the type age variable
has changed from a double (<dbl>) to character
(<chr>).
To simulate this situation:
Change the data type of the variable,
Tag the variable into a linelist, and then
Validate it.
Describe how linelist::validate_linelist() reacts when
there is a change in the data type of one variable of the input
data.
We can use dplyr::mutate() to change the variable type
before tagging for validation. For example:
R
# nolint start
cleaned_data %>%
# simulate a change of data type in one variable
dplyr::mutate(age = as.character(age)) %>%
# tag one variable
linelist::.... %>%
# validate the linelist
linelist::...
# nolint end
Please run the code line by line, focusing only on the parts before
the pipe (%>%). After each step, observe the output
before moving to the next line.
R
cleaned_data %>%
# simulate a change of data type in one variable
dplyr::mutate(age = as.character(age)) %>%
# tag one variable
linelist::make_linelist(age = "age") %>%
# validate the linelist
linelist::validate_linelist()
ERROR
Error:
! Some tags have the wrong class:
- age: Must inherit from class 'numeric'/'integer', but has class 'character'Challenge (continued)
Why are we getting an Error message?
Should we have a Warning message instead? Explain
why.
Explore other situations to understand this behavior by
converting:-date_onset from <date> to
character (<chr>), -gender character
(<chr>) to integer (<int>).
Then tag them into a linelist for validation. Does the
Error message suggest a fix to the issue?
Why are we getting an Error message? Should we have a
Warning message instead? Explain why? Explore other
situations to understand this behavior by
converting:-date_onset from <date> to
character (<chr>), -gender character
(<chr>) to integer (<int>).
Then tag them into a linelist for validation. Does the
Error message suggest a fix to the issue?
R
# Change 2
# Run this code line by line to identify changes
cleaned_data %>%
# simulate a change of data type
dplyr::mutate(date_onset = as.character(date_onset)) %>%
# tag
linelist::make_linelist(date_onset = "date_onset") %>%
# validate
linelist::validate_linelist()
R
# Change 3
# Run this code line by line to identify changes
cleaned_data %>%
# simulate a change of data type
dplyr::mutate(gender = as.factor(gender)) %>%
dplyr::mutate(gender = as.integer(gender)) %>%
# tag
linelist::make_linelist(gender = "gender") %>%
# validate
linelist::validate_linelist()
We get Error messages because the default type of these
variable in linelist::tags_types() is different from the
one we set them at.
The Error message inform us that in order to
validate our linelist, we must fix the input variable
type to fit the expected tag type. In a data analysis script, we can do
this by adding one cleaning step into the pipeline.
Challenge (continued)
Challenge
Beyond tagging and validating the linelist object, what extra step do we needed when building the object?
Let’s simulate a scenario about losing a variable :
R
cleaned_data %>%
# remove the variable 'age'
select(-age) %>%
# tag variable 'age' that no longer exist
linelist::make_linelist(
age = "age"
)
ERROR
Error in `base::tryCatch()`:
! 1 assertions failed:
* Variable 'tag': Must be element of set
* {'v1','case_id','gender','status','date_onset','date_sample','years_since_collection','remainder_months'},
* but is 'age'.Safeguarding
Safeguarding is implicitly built into the linelist objects. If you try to drop any of the tagged columns, you will receive an error or warning message, as shown in the example below.
R
new_df <- linelist_data %>%
dplyr::select(case_id, gender)
WARNING
Warning: The following tags have lost their variable:
date_onset:date_onsetThis Warning message above is the default output option
when we lose tags in a linelist object. However, it can be
changed to an Error message using the
linelist::lost_tags_action() function.
Challenge
Let’s test the implications of changing the
safeguarding configuration from a Warning
to an Error message.
- First, run this code to count the frequency of each category within a categorical variable:
R
linelist_data %>%
dplyr::select(case_id, gender) %>%
dplyr::count(gender)
- Set the behavior for lost tags in a
linelistto “error” as follows:
R
# set behavior to "error"
linelist::lost_tags_action(action = "error")
- Now, re - run the above code chunk with
dplyr::count().
Identify:
What is the difference in the output between a
Warningand anError?What could be the implications of this change for your daily data analysis pipeline during an outbreak response?
Deciding between Warning or Error message
will depend on the level of attention or flexibility you need when
losing tags. One will alert you about a change but will continue running
the code downstream. The other will stop your analysis pipeline and the
rest will not be executed.
A data reading, cleaning and validation script may require a more stable or fixed pipeline. An exploratory data analysis may require a more flexible approach. These two processes can be isolated in different scripts or repositories to adjust the safeguarding according to your needs.
Before you continue, set the configuration back again to the default
option of Warning:
R
# set behavior to the default option: "warning"
linelist::lost_tags_action()
OUTPUT
Lost tags will now issue a warning.A linelist object resembles a data frame but offers
richer features and functionalities. Packages that are linelist - aware
can leverage these features. For example, you can extract a data frame
of only the tagged columns using the linelist::tags_df()
function, as shown below:
R
linelist::tags_df(linelist_data)
OUTPUT
# A tibble: 15,000 × 3
id date_onset gender
<int> <IDate> <chr>
1 14905 2015-03-15 male
2 13043 2013-09-11 female
3 14364 2014-02-09 female
4 14675 2014-10-19 <NA>
5 12648 2014-06-08 female
6 14274 2015-04-05 female
7 14132 NA male
8 14715 NA female
9 13435 2014-07-09 male
10 14816 2015-06-29 female
# ℹ 14,990 more rowsThis allows for the use of tagged variables only in downstream analysis, which will be useful for the next episode!
When should I use
{linelist}?
Data analysis during an outbreak response or mass - gathering surveillance demands a different set of “data safeguards” if compared to usual research situations. For example, your data will change or be updated over time (e.g. new entries, new variables, renamed variables).
linelist is more appropriate for this type of ongoing
or long - lasting analysis. Check the “Get started” vignette section
about When
I should consider using {linelist}? for more
information.
Key Points
- Use the linelist package to tag, validate, and prepare case data for downstream analysis.
Content from Aggregate and visualize
Last updated on 2026-02-24 | Edit this page
Overview
Questions
- How to aggregate and summarise case data?
- How to visualize aggregated data?
- What is distribution of cases across time, space, gender, and age?
Objectives
- Simulate synthetic outbreak data
- Convert linelist data into incidence over time
- Create epidemic curves from incidence data
Introduction
In an analytic pipeline, exploratory data analysis (EDA) is an important step before formal modelling. EDA helps determine relationships between variables and summarize their main characteristics, often by means of data visualization.
This episode focuses on EDA of outbreak data using R packages. A key aspect of EDA in epidemic analysis is ‘person, place and time’. It is useful to identify how observed events - such as confirmed cases, hospitalizations, deaths, and recoveries - change over time, and how these vary across different locations and demographic factors, including gender, age, and more.
Let’s start by loading the incidence2 package to
aggregate the linelist data according to specific characteristics, and
visualize the resulting epidemic curves (epicurves) that plot the number
of new events (i.e. case incidence over time). We’ll use the
simulist package to simulate the outbreak data to
analyse, and {tracetheme} for figure formatting. We’ll use
the pipe operator (%>%) to connect some of their
functions, including others from the dplyr and
ggplot2 packages, so let’s also call to the {tidyverse}
package.
R
# Load packages
library(incidence2) # For aggregating and visualising
library(simulist) # For simulating linelist data
library(tracetheme) # For formatting figures
library(tidyverse) # For {dplyr} and {ggplot2} functions and the pipe |>
Synthetic outbreak data
To illustrate the process of conducting EDA on outbreak data, we will generate a line list for a hypothetical disease outbreak utilizing the simulist package. simulist generates simulated data for outbreak according to a given configuration. Its minimal configuration can generate a linelist, as shown in the below code chunk:
R
# Simulate linelist data for an outbreak with size between 1000 and 1500
set.seed(1) # Set seed for reproducibility
sim_data <- simulist::sim_linelist(outbreak_size = c(1000, 1500)) %>%
dplyr::as_tibble() # for a simple data frame output
WARNING
Warning: Number of cases exceeds maximum outbreak size.
Returning data early with 1546 cases and 3059 total contacts (including cases).R
# Display the simulated dataset
sim_data
OUTPUT
# A tibble: 1,546 × 13
id case_name case_type sex age date_onset date_reporting
<int> <chr> <chr> <chr> <int> <date> <date>
1 1 Travis Kurek confirmed m 37 2023-01-01 2023-01-01
2 3 Courtney Mccoy probable f 12 2023-01-11 2023-01-11
3 6 Andrea Alarid confirmed f 53 2023-01-18 2023-01-18
4 8 Salwa el-Sharifi suspected f 36 2023-01-23 2023-01-23
5 11 Azza al-Noorani suspected f 77 2023-01-30 2023-01-30
6 14 Olivya Pinto probable f 37 2023-01-24 2023-01-24
7 15 Acineth Briones suspected f 67 2023-01-31 2023-01-31
8 16 Mahuroos el-Javed confirmed m 80 2023-01-30 2023-01-30
9 20 Awad el-Idris probable m 70 2023-01-27 2023-01-27
10 21 Matthew Friend confirmed m 87 2023-02-09 2023-02-09
# ℹ 1,536 more rows
# ℹ 6 more variables: date_admission <date>, outcome <chr>,
# date_outcome <date>, date_first_contact <date>, date_last_contact <date>,
# ct_value <dbl>This linelist dataset has simulated entries on individual-level events during an outbreak.
The above is the default configuration of simulist. It
includes a number of assumptions about the transmissibility and severity
of the pathogen. If you want to know more about the
simulist::sim_linelist() function and other functionalities
check the documentation
website.
You can also find data sets from past real outbreaks within the {outbreaks}
R package.
Aggregating the data
Often we want to analyse and visualise the number of events that
occur on a particular day or week, rather than focusing on individual
cases. This requires grouping the linelist data into incidence data. The
{incidence2}
package offers a useful function called
incidence2::incidence() for grouping case data, usually
based around dated events and/or other characteristics. The code chunk
provided below demonstrates the creation of an
<incidence2> class object from the simulated Ebola
linelist data based on the date of onset.
R
# Create an incidence object by aggregating case data based on the date of onset
daily_incidence <- incidence2::incidence(
sim_data,
date_index = "date_onset",
interval = "day" # Aggregate by daily intervals
)
# View the incidence data
daily_incidence
OUTPUT
# incidence: 232 x 3
# count vars: date_onset
date_index count_variable count
<date> <chr> <int>
1 2023-01-01 date_onset 1
2 2023-01-11 date_onset 1
3 2023-01-18 date_onset 1
4 2023-01-23 date_onset 1
5 2023-01-24 date_onset 1
6 2023-01-27 date_onset 2
7 2023-01-29 date_onset 1
8 2023-01-30 date_onset 2
9 2023-01-31 date_onset 2
10 2023-02-01 date_onset 1
# ℹ 222 more rowsWith the incidence2 package, you can specify the desired interval (e.g. day, week) and categorize cases by one or more factors. Below is a code snippet demonstrating weekly cases grouped by the date of onset, sex, and type of case.
R
# Group incidence data by week, accounting for sex and case type
weekly_incidence <- incidence2::incidence(
sim_data,
date_index = "date_onset",
interval = "week", # Aggregate by weekly intervals
groups = c("sex", "case_type") # Group by sex and case type
)
# View the incidence data
weekly_incidence
OUTPUT
# incidence: 199 x 5
# count vars: date_onset
# groups: sex, case_type
date_index sex case_type count_variable count
<isowk> <chr> <chr> <chr> <int>
1 2022-W52 m confirmed date_onset 1
2 2023-W02 f probable date_onset 1
3 2023-W03 f confirmed date_onset 1
4 2023-W04 f probable date_onset 1
5 2023-W04 f suspected date_onset 1
6 2023-W04 m confirmed date_onset 1
7 2023-W04 m probable date_onset 2
8 2023-W05 f confirmed date_onset 5
9 2023-W05 f probable date_onset 2
10 2023-W05 f suspected date_onset 2
# ℹ 189 more rowsDates Completion
When cases are grouped by different factors, it’s possible that the
events involving these groups may have different date ranges in the
resulting incidence2 object. The incidence2
package provides a function called
incidence2::complete_dates() to ensure that an incidence
object has the same range of dates for each group. By default, missing
counts for a particular group will be filled with 0 for that date.
This functionality is also available within the
incidence2::incidence() function by setting the value of
the complete_dates to TRUE.
R
# Create a daily incidence object grouped by sex
daily_incidence_2 <- incidence2::incidence(
sim_data,
date_index = "date_onset",
groups = "sex",
interval = "day", # Aggregate by daily intervals
complete_dates = TRUE # Complete missing dates in the incidence object
)
Challenge 1: Can you do it?
-
Task: Calculate the biweekly
incidence of cases from the
sim_datalinelist based on their admission date and outcome. Save the result in an object calledbiweekly_incidence.
Visualization
The incidence2 objects can be visualized using the
plot() function from the base R package. The resulting
graph is referred to as an epidemic curve, or epi-curve for short. The
following code snippets generate epi-curves for the
daily_incidence and weekly_incidence incidence
objects mentioned above.
R
# Plot daily incidence data
base::plot(daily_incidence) +
ggplot2::labs(
x = "Time (in days)", # x-axis label
y = "Dialy cases" # y-axis label
) +
theme_bw()
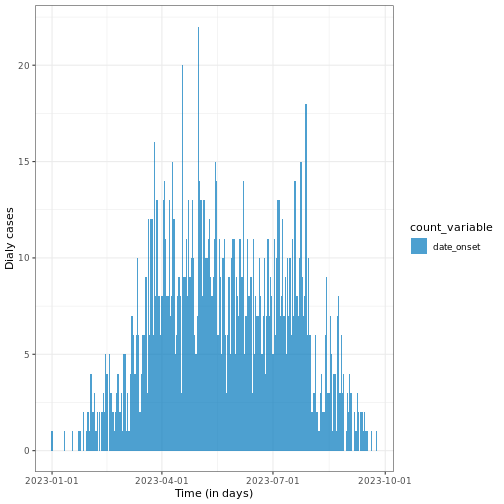
R
# Plot weekly incidence data
base::plot(weekly_incidence) +
ggplot2::labs(
x = "Time (in weeks)", # x-axis label
y = "weekly cases" # y-axis label
) +
theme_bw()
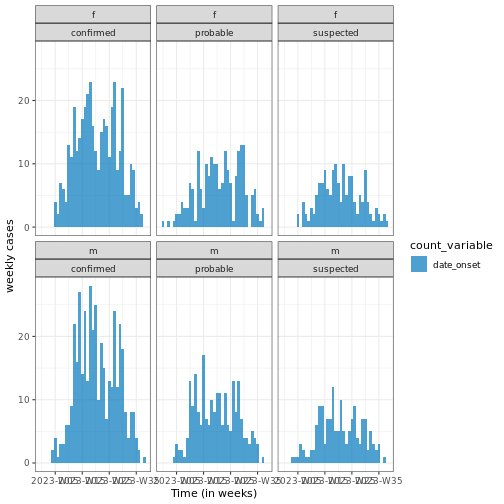
Easy aesthetics
We invite you to take a look at the incidence2 package
vignette. Find how you can use the arguments within the
plot() function to provide aesthetics to your incidence2
class objects.
R
base::plot(weekly_incidence, fill = "sex")
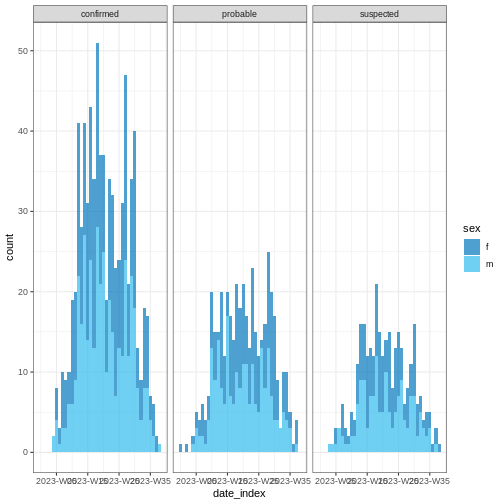
Some of them include show_cases = TRUE,
angle = 45, and n_breaks = 5. Try them and see
how they impact on the resulting plot.
Challenge 2: Can you do it?
-
Task: Visualize the
biweekly_incidenceobject.
Curve of cumulative cases
The cumulative number of cases can be calculated using the
incidence2::cumulate() function on an
incidence2 object and visualized it, as in the example
below.
R
# Calculate cumulative incidence
cum_df <- incidence2::cumulate(daily_incidence)
# Plot cumulative incidence data using ggplot2
base::plot(cum_df) +
ggplot2::labs(
x = "Time (in days)", # x-axis label
y = "weekly cases" # y-axis label
) +
theme_bw()
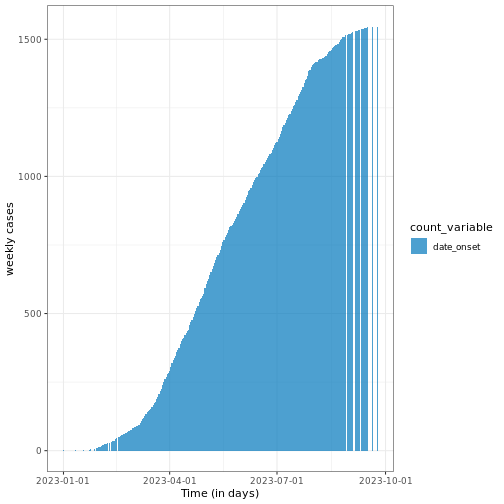
Note that this function preserves grouping, i.e., if the
incidence2 object contains groups, it will accumulate the
cases accordingly.
Challenge 3: Can you do it?
-
Task: Visulaize the cumulative cases from the
biweekly_incidenceobject.
Peak time estimation
You can estimate the peak – the time with the highest number of
recorded cases – using the incidence2::estimate_peak()
function from the {incidence2} package. This function uses a
bootstrapping method to determine the peak time (i.e. by resampling
dates with replacement, resulting in a distribution of estimated peak
times).
R
# Estimate the peak of the daily incidence data
peak <- incidence2::estimate_peak(
daily_incidence,
n = 100, # Number of simulations for the peak estimation
alpha = 0.05, # Significance level for the confidence interval
first_only = TRUE, # Return only the first peak found
progress = FALSE # Disable progress messages
)
# Display the estimated peak
print(peak)
OUTPUT
# A tibble: 1 × 7
count_variable observed_peak observed_count bootstrap_peaks lower_ci
<chr> <date> <int> <list> <date>
1 date_onset 2023-05-01 22 <df [100 × 1]> 2023-03-26
# ℹ 2 more variables: median <date>, upper_ci <date>This example demonstrates how to estimate the peak time using the
incidence2::estimate_peak() function at \(95%\) confidence interval and using 100
bootstrap samples.
Challenge 4: Can you do it?
-
Task: Estimate the peak time from the
biweekly_incidenceobject.
Visualization with ggplot2
incidence2 produces basic plots for epicurves, but additional work is required to create well-annotated graphs. However, using the ggplot2 package, you can generate more sophisticated epicurves, with more flexibility in annotation. ggplot2 is a comprehensive package with many functionalities. However, we will focus on three key elements for producing epicurves: histogram plots, scaling date axes and their labels, and general plot theme annotation. The example below demonstrates how to configure these three elements for a simple incidence2 object.
R
# Define date breaks for the x-axis
breaks <- seq.Date(
from = min(as.Date(daily_incidence$date_index, na.rm = TRUE)),
to = max(as.Date(daily_incidence$date_index, na.rm = TRUE)),
by = 20 # every 20 days
)
# Create the plot
ggplot2::ggplot(data = daily_incidence) +
geom_histogram(
mapping = aes(
x = as.Date(date_index),
y = count
),
stat = "identity",
color = "blue", # bar border color
fill = "lightblue", # bar fill color
width = 1 # bar width
) +
theme_minimal() + # apply a minimal theme for clean visuals
theme(
plot.title = element_text(face = "bold",
hjust = 0.5), # center and bold title
plot.subtitle = element_text(hjust = 0.5), # center subtitle
plot.caption = element_text(face = "italic",
hjust = 0), # italicized caption
axis.title = element_text(face = "bold"), # bold axis titles
axis.text.x = element_text(angle = 45, vjust = 0.5) # rotated x-axis text
) +
labs(
x = "Date", # x-axis label
y = "Number of cases", # y-axis label
title = "Daily Outbreak Cases", # plot title
subtitle = "Epidemiological Data for the Outbreak", # plot subtitle
caption = "Data Source: Simulated Data" # plot caption
) +
scale_x_date(
breaks = breaks, # set custom breaks on the x-axis
labels = scales::label_date_short() # shortened date labels
)
WARNING
Warning in geom_histogram(mapping = aes(x = as.Date(date_index), y = count), :
Ignoring unknown parameters: `binwidth` and `bins`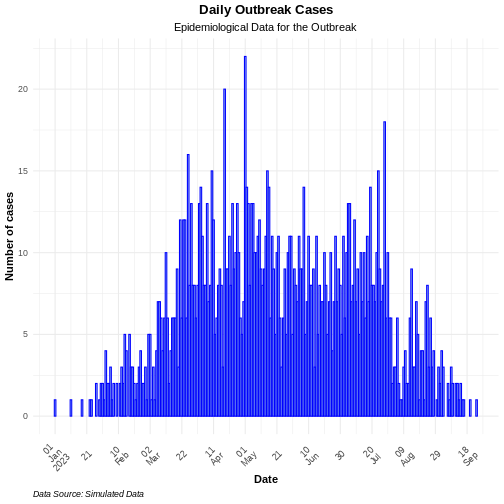
Use the group option in the mapping function to
visualize an epicurve with different groups. If there is more than one
grouping factor, use the facet_wrap() option, as
demonstrated in the example below:
R
# Plot daily incidence faceted by sex
ggplot2::ggplot(data = daily_incidence_2) +
geom_histogram(
mapping = aes(
x = as.Date(date_index),
y = count,
group = sex,
fill = sex
),
stat = "identity"
) +
theme_minimal() + # apply minimal theme
theme(
plot.title = element_text(face = "bold",
hjust = 0.5), # bold and center the title
plot.subtitle = element_text(hjust = 0.5), # center the subtitle
plot.caption = element_text(face = "italic", hjust = 0), # italic caption
axis.title = element_text(face = "bold"), # bold axis labels
axis.text.x = element_text(angle = 45,
vjust = 0.5) # rotate x-axis text for readability
) +
labs(
x = "Date", # x-axis label
y = "Number of cases", # y-axis label
title = "Daily Outbreak Cases by Sex", # plot title
subtitle = "Incidence of Cases Grouped by Sex", # plot subtitle
caption = "Data Source: Simulated Data" # caption for additional context
) +
facet_wrap(~sex) + # create separate panels by sex
scale_x_date(
breaks = breaks, # set custom date breaks
labels = scales::label_date_short() # short date format for x-axis labels
) +
scale_fill_manual(values = c("lightblue",
"lightpink")) # custom fill colors for sex
WARNING
Warning in geom_histogram(mapping = aes(x = as.Date(date_index), y = count, :
Ignoring unknown parameters: `binwidth` and `bins`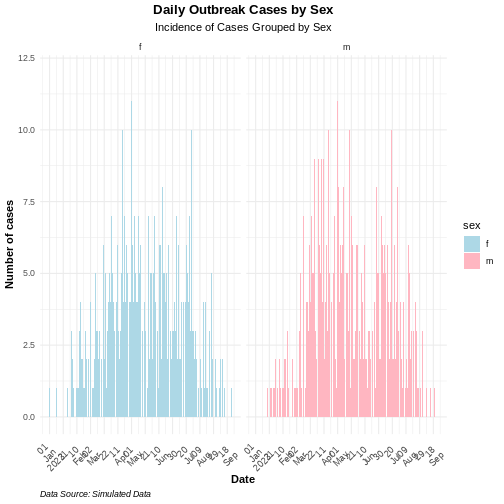
Challenge 5: Can you do it?
-
Task: Produce an annotated figure for the
biweekly_incidenceobject using the ggplot2 package.
Key Points
- Use simulist package to generate synthetic outbreak data
- Use incidence2 package to aggregate case data based on a date event, and other variables to produce epidemic curves.
- Use ggplot2 package to produce better annotated epicurves.
