Clean case data
Last updated on 2026-02-24 | Edit this page
Estimated time: 40 minutes
Overview
Questions
- How to clean and standardize case data?
Objectives
- Explain how to clean, curate, and standardize case data using cleanepi package
- Perform essential data-cleaning operations on a real case dataset.
Prerequisite
In this episode, we will use a simulated Ebola dataset that can be:
- Download the simulated_ebola_2.csv
- Save it in the
data/folder. Follow instructions in Setup to configure an RStudio Project and folder
Introduction
In the process of analyzing outbreak data, it’s essential to ensure that the dataset is clean, curated, standardized, and validated. This will ensure that analysis is accurate (i.e. you are analysing what you think you are analysing) and reproducible (i.e. if someone wants to go back and repeat your analysis steps with your code, you can be confident they will get the same results). This episode focuses on cleaning epidemics and outbreaks data using the cleanepi package, For demonstration purposes, we’ll work with a simulated dataset of Ebola cases.
Let’s start by loading the package rio to read data
and the package cleanepi to clean it. We’ll use the pipe
%>% to connect some of their functions, including others
from the package dplyr, so let’s also call to the
tidyverse package:
R
# Load packages
library(tidyverse) # for {dplyr} functions and the pipe %>%
library(rio) # for importing data
library(here) # for easy file referencing
library(cleanepi)
The double-colon (::)
operator
The :: in R lets you access functions or objects from a
specific package without attaching the entire package to the search
path. It offers several important advantages including the
followings:
- Telling explicitly which package a function comes from, reducing ambiguity and potential conflicts when several packages have functions with the same name.
- Allowing to call a function from a package without loading the whole package with library().
For example, the command dplyr::filter(data, condition)
means we are calling the filter() function from the
dplyr package.
The first step is to import the dataset into working environment.
This can be done by following the guidelines outlined in the Read case data episode. It involves loading
the dataset into R environment and view its structure and
content.
R
# Read data
# e.g.: if path to file is data/simulated_ebola_2.csv then:
raw_ebola_data <- rio::import(
here::here("data", "simulated_ebola_2.csv")
) %>%
dplyr::as_tibble() # for a simple data frame output
R
# Print data frame
raw_ebola_data
OUTPUT
# A tibble: 15,003 × 9
V1 `case id` age gender status `date onset` `date sample` lab region
<int> <int> <chr> <chr> <chr> <chr> <chr> <lgl> <chr>
1 1 14905 90 1 "conf… 03/15/2015 06/04/2015 NA valdr…
2 2 13043 twenty… 2 "" Sep /11/13 03/01/2014 NA valdr…
3 3 14364 54 f <NA> 09/02/2014 03/03/2015 NA valdr…
4 4 14675 ninety <NA> "" 10/19/2014 31/ 12 /14 NA valdr…
5 5 12648 74 F "" 08/06/2014 10/10/2016 NA valdr…
6 5 12648 74 F "" 08/06/2014 10/10/2016 NA valdr…
7 6 14274 sevent… female "" Apr /05/15 01/23/2016 NA valdr…
8 7 14132 sixteen male "conf… Dec /29/Y 05/10/2015 NA valdr…
9 8 14715 44 f "conf… Apr /06/Y 04/24/2016 NA valdr…
10 9 13435 26 1 "" 09/07/2014 20/ 09 /14 NA valdr…
# ℹ 14,993 more rowsDiscussion
Let’s first diagnose the data frame. List all the characteristics in the data frame above that are problematic for data analysis.
Are any of those characteristics familiar from any previous data analysis you have performed?
Lead a short discussion to relate the diagnosed characteristics with required cleaning operations.
You can use the following terms to diagnose characteristics:
- Codification, like the codification of values in columns like ‘gender’ and ‘age’ using numbers, letters, and words. Also the presence of multiple dates formats (“dd/mm/yyyy”, “yyyy/mm/dd”, etc) in the same column like in ‘date_onset’. Less visible, but also the column names.
- Missing, how to interpret an entry like “” in the ‘status’ column or “-99” in other circumstances? Do we have a data dictionary from the data collection process?
- Inconsistencies, like having a date of sample before the date of onset.
- Non-plausible values, like observations where some dates values are outside of the expected timeframe.
- Duplicates, are all observations unique?
You can use these terms to relate to cleaning operations:
- Standardize column name
- Standardize categorical variables like ‘gender’
- Standardize date columns
- Convert character values into numeric
- Check the sequence of dated events
A quick inspection
Quick exploration and inspection of the dataset are crucial to
identify potential data issues before diving into any analysis tasks.
The cleanepi package simplifies this process with the
scan_data() function. Let’s take a look at how you can use
it:
R
cleanepi::scan_data(raw_ebola_data)
OUTPUT
Field_names missing numeric date character logical
1 age 0.0690 0.8925 0.0000 0.1075 0
2 gender 0.1874 0.0560 0.0000 0.9440 0
3 status 0.0565 0.0000 0.0000 1.0000 0
4 date onset 0.0001 0.0000 0.9159 0.0841 0
5 date sample 0.0001 0.0000 1.0000 0.0000 0
6 region 0.0000 0.0000 0.0000 1.0000 0The results provide an overview of the content of all character columns, including column names, and the percent of some data types within them. You can see that the column names in the dataset are descriptive but lack consistency. Some are composed of multiple words separated by white spaces. Additionally, some columns contain more than one data type, and there are missing values in the form of an empty string in others.
Common operations
This section demonstrate how to perform some common data cleaning operations using the cleanepi package.
Standardizing column names
For this example dataset, standardizing column names typically
involves removing with spaces and connecting different words with “_”.
This practice helps maintain consistency and readability in the dataset.
However, the function used for standardizing column names offers more
options. Type ?cleanepi::standardize_column_names in the
console for more details.
R
sim_ebola_data <- cleanepi::standardize_column_names(raw_ebola_data)
names(sim_ebola_data)
OUTPUT
[1] "v1" "case_id" "age" "gender" "status"
[6] "date_onset" "date_sample" "lab" "region" If you want to maintain certain column names without subjecting them
to the standardization process, you can utilize the keep
argument of the function
cleanepi::standardize_column_names(). This argument accepts
a vector of column names that are intended to be kept unchanged.
Challenge
What differences can you observe in the column names?
Standardize the column names of the input dataset, but keep the first column names as it is.
You can try
cleanepi::standardize_column_names(data = raw_ebola_data, keep = "V1")
Removing irregularities
Raw data may contain fields that don’t add any variability to the
data such as empty rows and columns, or
constant columns (where all entries have the same
value). It can also contain duplicated rows. Functions
from cleanepi like remove_duplicates() and
remove_constants() remove such irregularities as
demonstrated in the below code chunk.
R
# Remove constants
sim_ebola_data <- cleanepi::remove_constants(sim_ebola_data)
Now, print the output to identify what constant column you removed!
R
# Remove duplicates
sim_ebola_data <- cleanepi::remove_duplicates(sim_ebola_data)
OUTPUT
! Found 5 duplicated rows in the dataset.
ℹ Use `print_report(dat, "found_duplicates")` to access them, where "dat" is
the object used to store the output from this operation.You can get the number and location of the duplicated rows that where
found. Run cleanepi::print_report(), wait for the report to
open in your browser, and find the “Duplicates” tab.
To use this information within R, you can print data frames with
specific sections of the report in the console using the argument
what.
R
# Print a report of found duplicates
cleanepi::print_report(data = sim_ebola_data, what = "found_duplicates")
# Print a report of removed duplicates
cleanepi::print_report(data = sim_ebola_data, what = "removed_duplicates")
Challenge
In the following data frame:
OUTPUT
# A tibble: 6 × 5
col1 col2 col3 col4 col5
<dbl> <dbl> <chr> <chr> <date>
1 1 1 a b NA
2 2 3 a b NA
3 NA NA a <NA> NA
4 NA NA a <NA> NA
5 NA NA a <NA> NA
6 NA NA <NA> <NA> NA What columns are the:
- constant data?
- duplicated rows?
Constant data mostly refers to empty rows or columns as well as constant columns.
Make sure they start by removing duplicates before removing constant data.
- indices of duplicated rows: 3, 4, 5
- indices of empty rows: 4 (from the first iteration); 3 (from the second iteration)
- empty cols: “col5”
- constant cols: “col3”, and “col4”
Point out to learners that they create a different set of constant
data after removing by varying the value of the cutoff
argument.
R
df <- df %>% cleanepi::remove_constants(cutoff = 0.5)
Replacing missing values
In addition to the irregularities, raw data may contain missing
values, and these may be encoded by different strings
(e.g. "NA", "", character(0)). To
ensure robust analysis, it is a good practice to replace all missing
values by NA in the entire dataset. Below is a code snippet
demonstrating how you can achieve this in cleanepi for
missing entries represented by an empty string "":
R
sim_ebola_data <- cleanepi::replace_missing_values(
data = sim_ebola_data,
na_strings = ""
)
sim_ebola_data
OUTPUT
# A tibble: 15,000 × 7
v1 case_id age gender status date_onset date_sample
<int> <int> <chr> <chr> <chr> <chr> <chr>
1 1 14905 90 1 confirmed 03/15/2015 06/04/2015
2 2 13043 twenty-five 2 <NA> sep /11/13 03/01/2014
3 3 14364 54 f <NA> 09/02/2014 03/03/2015
4 4 14675 ninety <NA> <NA> 10/19/2014 31/ 12 /14
5 5 12648 74 F <NA> 08/06/2014 10/10/2016
6 6 14274 seventy-six female <NA> apr /05/15 01/23/2016
7 7 14132 sixteen male confirmed dec /29/y 05/10/2015
8 8 14715 44 f confirmed apr /06/y 04/24/2016
9 9 13435 26 1 <NA> 09/07/2014 20/ 09 /14
10 10 14816 thirty f <NA> 06/29/2015 06/02/2015
# ℹ 14,990 more rowsValidating subject IDs
Each entry in the dataset represents a subject (e.g. a disease case
or study participant) and should be distinguishable by a specific ID
formatted in a particular way. These IDs can contain numbers falling
within a specific range, a prefix and/or suffix, and might be written
such that they contain a specific number of characters. The
cleanepi package offers the function
check_subject_ids() designed precisely for this task as
shown in the below code chunk. This function checks whether the IDs are
unique and meet the required criteria specified by the user.
R
# check if the subject IDs in the 'case_id' column contains numbers ranging
# from 0 to 15000
sim_ebola_data <- cleanepi::check_subject_ids(
data = sim_ebola_data,
target_columns = "case_id",
range = c(0, 15000)
)
OUTPUT
! Detected 0 missing, 1957 duplicated, and 0 incorrect subject IDs.
ℹ Enter `print_report(data = dat, "incorrect_subject_id")` to access them,
where "dat" is the object used to store the output from this operation.
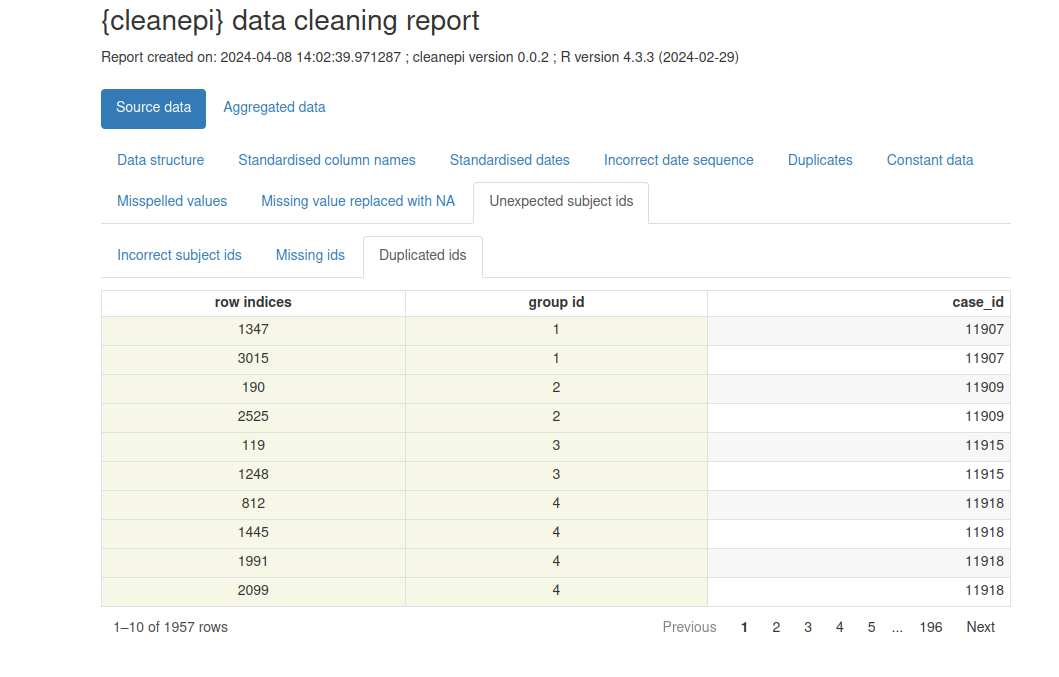
ℹ You can use the `correct_subject_ids()` function to correct them.Note that our simulated dataset contains duplicated subject IDs.
Let’s print a preliminary report with
cleanepi::print_report(sim_ebola_data). Focus on the
“Unexpected subject ids” tab to identify what IDs require an extra
treatment.
In the console, you can print:
R
print_report(data = sim_ebola_data, "incorrect_subject_id")
After finishing this tutorial, we invite you to explore the package reference guide of cleanepi::check_subject_ids() to find the function that can fix this situation.
Standardizing dates
An epidemic dataset typically contains date columns for different events, such as the date of infection, date of symptoms onset, etc. These dates can come in different date formats, and it is good practice to standardize them to benefit from the powerful R functionalities designed to handle date values in downstream analyses. The cleanepi package provides functionality for converting date columns of epidemic datasets into ISO8601 format, ensuring consistency across the different date columns. Here’s how you can use it on our simulated dataset:
R
sim_ebola_data <- cleanepi::standardize_dates(
sim_ebola_data,
target_columns = c("date_onset", "date_sample")
)
OUTPUT
! Detected 1142 values that comply with multiple formats and no values that are
outside of the specified time frame.
ℹ Enter `print_report(data = dat, "date_standardization")` to access them,
where "dat" is the object used to store the output from this operation.R
sim_ebola_data
OUTPUT
# A tibble: 15,000 × 7
v1 case_id age gender status date_onset date_sample
<int> <chr> <chr> <chr> <chr> <date> <date>
1 1 14905 90 1 confirmed 2015-03-15 2015-04-06
2 2 13043 twenty-five 2 <NA> 2013-09-11 2014-01-03
3 3 14364 54 f <NA> 2014-02-09 2015-03-03
4 4 14675 ninety <NA> <NA> 2014-10-19 2014-12-31
5 5 12648 74 F <NA> 2014-06-08 2016-10-10
6 6 14274 seventy-six female <NA> 2015-04-05 2016-01-23
7 7 14132 sixteen male confirmed NA 2015-10-05
8 8 14715 44 f confirmed NA 2016-04-24
9 9 13435 26 1 <NA> 2014-07-09 2014-09-20
10 10 14816 thirty f <NA> 2015-06-29 2015-02-06
# ℹ 14,990 more rowsThis function converts the values in the target columns into the YYYY-mm-dd format.
How is this possible?
We invite you to find the key package that makes this standardisation possible inside cleanepi by reading the “Details” section of the Standardize date variables reference manual!
Also, check how to use the orders argument if you want
to target US format character strings. You can explore this
reproducible example.
Converting to numeric values
In the raw dataset, some columns can come with mixture of character
and numerical values, and you will often want to convert character
values for numbers explicitly into numeric values
(e.g. "seven" to 7). For example, in our
simulated data set, in the age column some entries are written in words.
In cleanepi the function
convert_to_numeric() does such conversion as illustrated in
the below code chunk.
R
sim_ebola_data <- cleanepi::convert_to_numeric(
data = sim_ebola_data,
target_columns = "age"
)
sim_ebola_data
OUTPUT
# A tibble: 15,000 × 7
v1 case_id age gender status date_onset date_sample
<int> <chr> <dbl> <chr> <chr> <date> <date>
1 1 14905 90 1 confirmed 2015-03-15 2015-04-06
2 2 13043 25 2 <NA> 2013-09-11 2014-01-03
3 3 14364 54 f <NA> 2014-02-09 2015-03-03
4 4 14675 90 <NA> <NA> 2014-10-19 2014-12-31
5 5 12648 74 F <NA> 2014-06-08 2016-10-10
6 6 14274 76 female <NA> 2015-04-05 2016-01-23
7 7 14132 16 male confirmed NA 2015-10-05
8 8 14715 44 f confirmed NA 2016-04-24
9 9 13435 26 1 <NA> 2014-07-09 2014-09-20
10 10 14816 30 f <NA> 2015-06-29 2015-02-06
# ℹ 14,990 more rowsMultiple language support
Thanks to the numberize package, we can convert numbers written in English, French or Spanish into positive integer values.
Epidemiology related operations
In addition to common data cleansing tasks, such as those discussed in the above section, the cleanepi package offers additional functionalities tailored specifically for processing and analyzing outbreak and epidemic data. This section covers some of these specialized tasks.
Checking sequence of dated-events
Ensuring the correct order and sequence of dated events is crucial in
epidemiological data analysis, especially when analyzing infectious
diseases where the timing of events like symptom onset and sample
collection is essential. The cleanepi package provides a
helpful function called check_date_sequence() designed for
this purpose.
Here’s an example of a code chunk demonstrating the usage of the
function check_date_sequence() in the first 100 records of
our simulated Ebola dataset.
R
cleanepi::check_date_sequence(
data = sim_ebola_data[1:100, ],
target_columns = c("date_onset", "date_sample")
)
OUTPUT
ℹ Cannot check the sequence of date events across 37 rows due to missing data.OUTPUT
! Detected 24 incorrect date sequences at lines: "8, 15, 18, 20, 21, 23, 26,
28, 29, 32, 34, 35, 37, 38, 40, 43, 46, 49, 52, 54, 56, 58, 60, 63".
ℹ Enter `print_report(data = dat, "incorrect_date_sequence")` to access them,
where "dat" is the object used to store the output from this operation.This functionality is crucial for ensuring data integrity and accuracy in epidemiological analyses, as it helps identify any inconsistencies or errors in the chronological order of events, allowing you to address them appropriately.
Dictionary-based substitution
In the realm of data pre-processing, it’s common to encounter scenarios where certain columns in a dataset, such as the “gender” column in our simulated Ebola dataset, are expected to have specific values or factors. However, it’s also common for unexpected or erroneous values to appear in these columns, which need to be replaced with the appropriate values. The cleanepi package offers support for dictionary-based substitution, a method that allows you to replace values in specific columns based on mappings defined in a data dictionary. This approach ensures consistency and accuracy in data cleaning.
Moreover, cleanepi provides a built-in dictionary specifically tailored for epidemiological data. The example dictionary below includes mappings for the “gender” column.
R
test_dict <- base::readRDS(
system.file("extdata", "test_dict.RDS", package = "cleanepi")
) %>%
dplyr::as_tibble()
test_dict
OUTPUT
# A tibble: 6 × 4
options values grp orders
<chr> <chr> <chr> <int>
1 1 male gender 1
2 2 female gender 2
3 M male gender 3
4 F female gender 4
5 m male gender 5
6 f female gender 6Now, we can use this dictionary to standardize values of the the
“gender” column according to predefined categories. Below is an example
code chunk demonstrating how to perform this using the
clean_using_dictionary() function from the {cleanepi}
package.
R
sim_ebola_data <- cleanepi::clean_using_dictionary(
data = sim_ebola_data,
dictionary = test_dict
)
sim_ebola_data
OUTPUT
# A tibble: 15,000 × 7
v1 case_id age gender status date_onset date_sample
<int> <chr> <dbl> <chr> <chr> <date> <date>
1 1 14905 90 male confirmed 2015-03-15 2015-04-06
2 2 13043 25 female <NA> 2013-09-11 2014-01-03
3 3 14364 54 female <NA> 2014-02-09 2015-03-03
4 4 14675 90 <NA> <NA> 2014-10-19 2014-12-31
5 5 12648 74 female <NA> 2014-06-08 2016-10-10
6 6 14274 76 female <NA> 2015-04-05 2016-01-23
7 7 14132 16 male confirmed NA 2015-10-05
8 8 14715 44 female confirmed NA 2016-04-24
9 9 13435 26 male <NA> 2014-07-09 2014-09-20
10 10 14816 30 female <NA> 2015-06-29 2015-02-06
# ℹ 14,990 more rowsThis approach simplifies the data cleaning process, ensuring that categorical variables in epidemiological datasets are accurately categorized and ready for further analysis.
Note that, when a column in the dataset contains values that are not
in the dictionary, the function
cleanepi::clean_using_dictionary() will raise an error. You
can start a custom dictionary with a data frame inside or outside R and
use the function cleanepi::add_to_dictionary() to include
new elements in the dictionary. For example:
R
new_dictionary <- tibble::tibble(
options = "0",
values = "female",
grp = "sex",
orders = 1L
) %>%
cleanepi::add_to_dictionary(
option = "1",
value = "male",
grp = "sex",
order = NULL
)
new_dictionary
OUTPUT
# A tibble: 2 × 4
options values grp orders
<chr> <chr> <chr> <int>
1 0 female sex 1
2 1 male sex 2You can have more details in the section about “Dictionary-based data substituting” in the package vignette.
Calculating time span between different date events
In epidemiological data analysis, it is also useful to track and analyze time-dependent events, such as the progression of a disease outbreak (i.e., the time elapsed between today and the date the first case was reported) or the duration between date of sample collection and analysis (i.e., the time difference between today and the sample collection date). The most common example is to calculate the age of all the subjects given their dates of birth (i.e., the time difference between today and their date of birth).
The cleanepi package offers a convenient function for
calculating the time elapsed between two dated events at different time
scales. For example, the below code snippet utilizes the function
cleanepi::timespan() to compute the time elapsed since the
date of sampling of the identified cases until the \(3^{rd}\) of January 2025
("2025-01-03").
R
sim_ebola_data <- cleanepi::timespan(
data = sim_ebola_data,
target_column = "date_sample",
end_date = as.Date("2025-01-03"),
span_unit = "years",
span_column_name = "years_since_collection",
span_remainder_unit = "months"
)
sim_ebola_data %>%
dplyr::select(case_id, date_sample, years_since_collection, remainder_months)
OUTPUT
# A tibble: 15,000 × 4
case_id date_sample years_since_collection remainder_months
<chr> <date> <dbl> <dbl>
1 14905 2015-04-06 9 8
2 13043 2014-01-03 11 0
3 14364 2015-03-03 9 10
4 14675 2014-12-31 10 0
5 12648 2016-10-10 8 2
6 14274 2016-01-23 8 11
7 14132 2015-10-05 9 2
8 14715 2016-04-24 8 8
9 13435 2014-09-20 10 3
10 14816 2015-02-06 9 10
# ℹ 14,990 more rowsAfter executing the function cleanepi::timespan(), two
new columns named years_since_collection and
remainder_months are added to the
sim_ebola_data dataset. For each case, these columns
respectively represent the calculated time elapsed since the date of
sample collection measured in years, and the remaining time measured in
months.
Challenge
Age data is useful in many downstream analysis. You can categorize it to generate stratified estimates.
Read the test_df.RDS data frame within the
cleanepi package:
R
dat <- readRDS(
file = system.file("extdata", "test_df.RDS", package = "cleanepi")
) %>%
dplyr::as_tibble()
Calculate the age in years until today’s date of the subjects from their date of birth, and the remainder time in months. Clean and standardize the required elements to get this done.
Before calculating the age, you may need to:
- standardize column names
- standardize dates columns
- replace missing value strings with NA
In the solution we added date_first_pcr_positive_test as
part of the Date columns to be standardised given that it often used in
disease outbreak analysis.
R
dat_clean <- dat %>%
# standardize column names and dates
cleanepi::standardize_column_names() %>%
cleanepi::standardize_dates(
target_columns = c("date_of_birth", "date_first_pcr_positive_test")
) %>%
# replace missing value strings with NA
cleanepi::replace_missing_values(
target_columns = c("sex", "date_of_birth"),
na_strings = "-99"
) %>%
# calculate the age in 'years' and return the remainder in 'months'
cleanepi::timespan(
target_column = "date_of_birth",
end_date = Sys.Date(),
span_unit = "years",
span_column_name = "age_in_years",
span_remainder_unit = "months"
)
OUTPUT
! Detected 4 values that comply with multiple formats and no values that are
outside of the specified time frame.
ℹ Enter `print_report(data = dat, "date_standardization")` to access them,
where "dat" is the object used to store the output from this operation.
! Found <numeric> values that could also be of type <Date> in column:
date_of_birth.
ℹ It is possible to convert them into <Date> using: `lubridate::as_date(x,
origin = as.Date("1900-01-01"))`
• where "x" represents here the vector of values from these columns
(`data$target_column`).Now, How would you categorize a numerical variable?
The simplest alternative is using Hmisc::cut2(). You can
also use dplyr::case_when(). However, this requires more
lines of code and is more appropriate for custom categorization. Here we
provide one solution using base::cut():
R
dat_clean <- dat_clean %>%
# select the columns of interest
dplyr::select(
study_id,
sex,
date_first_pcr_positive_test,
date_of_birth,
age_in_years
) %>%
# categorize the age variable [add as a challenge hint]
dplyr::mutate(
age_category = base::cut(
x = age_in_years,
breaks = c(0, 20, 35, 60, Inf), # replace with max value if known
include.lowest = TRUE,
right = FALSE
)
)
You can investigate the maximum values of variables from the summary
made from the skimr::skim() function. Instead of
base::cut() you can also use
Hmisc::cut2(x = age_in_years, cuts = c(20,35,60)), which
gives the maximum value and do not require more arguments.
Multiple operations at once
Performing data cleaning operations individually can be
time-consuming and error-prone. The cleanepi package
simplifies this process by offering a convenient wrapper function called
clean_data(), which allows you to perform multiple
operations at once.
When no cleaning operation is specified, the
clean_data() function automatically applies a series of
data cleaning operations to the input dataset. Here’s an example code
chunk illustrating how to use clean_data() on a raw
simulated Ebola dataset:
R
cleaned_data <- cleanepi::clean_data(raw_ebola_data)
OUTPUT
ℹ Cleaning column namesOUTPUT
ℹ Removing constant columns and empty rowsOUTPUT
ℹ Removing duplicated rowsOUTPUT
! Found 5 duplicated rows in the dataset.
ℹ Use `print_report(dat, "found_duplicates")` to access them, where "dat" is
the object used to store the output from this operation.Further more, you can combine multiple data cleaning tasks via the
base R pipe (%>%) or the {magrittr} pipe
(%>%) operator, as shown in the below code snippet.
R
# Perform the cleaning operations using the pipe (%>%) operator
cleaned_data <- raw_ebola_data %>%
cleanepi::standardize_column_names() %>%
cleanepi::remove_constants() %>%
cleanepi::remove_duplicates() %>%
cleanepi::replace_missing_values(na_strings = "") %>%
cleanepi::check_subject_ids(
target_columns = "case_id",
range = c(1, 15000)
) %>%
cleanepi::standardize_dates(
target_columns = c("date_onset", "date_sample")
) %>%
cleanepi::convert_to_numeric(target_columns = "age") %>%
cleanepi::check_date_sequence(
target_columns = c("date_onset", "date_sample")
) %>%
cleanepi::clean_using_dictionary(dictionary = test_dict) %>%
cleanepi::timespan(
target_column = "date_sample",
end_date = as.Date("2025-01-03"),
span_unit = "years",
span_column_name = "years_since_collection",
span_remainder_unit = "months"
)
Challenge
Have you noticed that cleanepi contains a set of functions to diagnose the cleaning status of the dataset and another set to perform cleaning actions on it?
To identify both groups:
- On a piece of paper, write the names of each function under the corresponding column:
| Diagnose cleaning status | Perform cleaning action |
|---|---|
| … | … |
Notice that cleanepi contains a set of functions to
diagnose the cleaning status (e.g.,
check_subject_ids() and check_date_sequence()
in the chunk above) and another set to perform a
cleaning action (the complementary functions from the chunk above).
Cleaning report
The cleanepi package generates a comprehensive report detailing the findings and actions of all data cleansing operations conducted during the analysis. This report is presented as a HTML file that automatically opens in your browser with. Each section corresponds to a specific data cleansing operation, and clicking on each section allows you to access the results of that particular operation. This interactive approach enables users to efficiently review and analyze the effects of individual cleansing steps within the broader data cleansing process.
You can view the report using:
R
cleanepi::print_report(data = cleaned_data)
Example of data cleaning report generated by cleanepi