Read case data
Clean case data
Figure 1
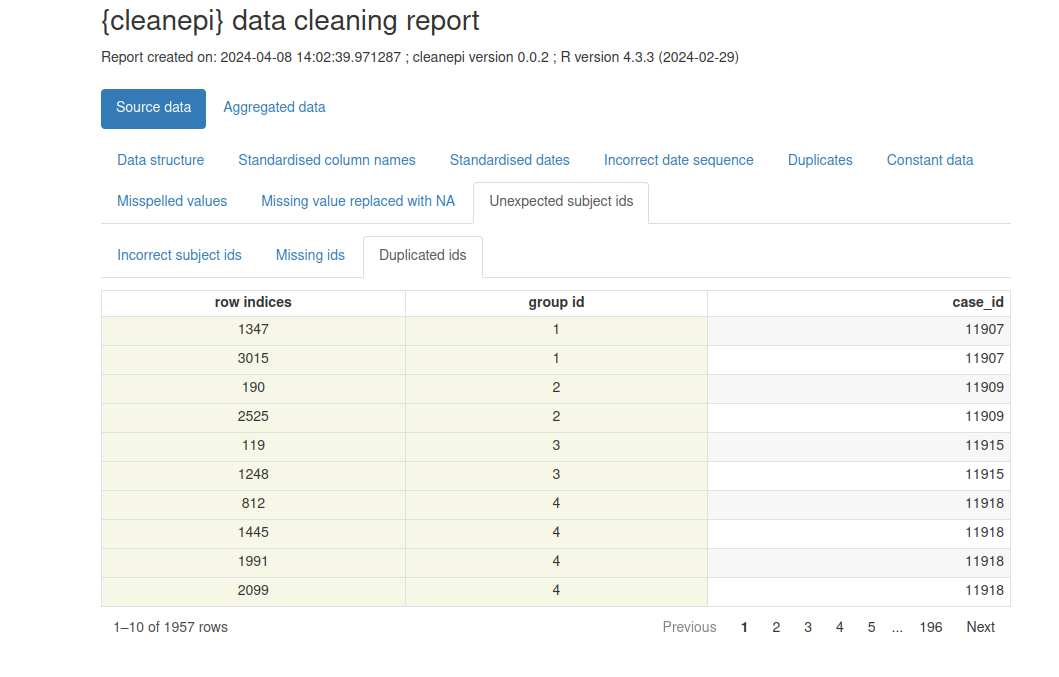
Example of data cleaning report generated by cleanepi
Validate case data
Aggregate and visualize
Figure 1
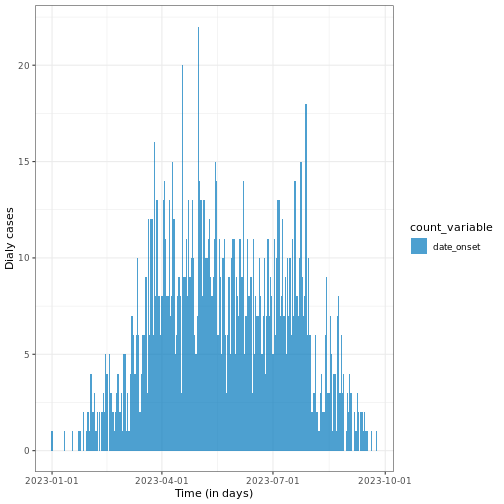
Figure 2
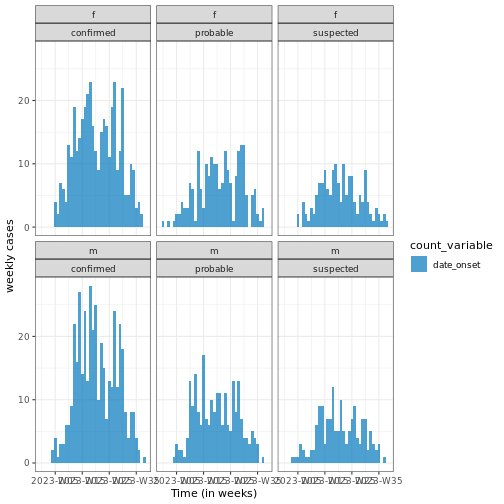
Figure 3
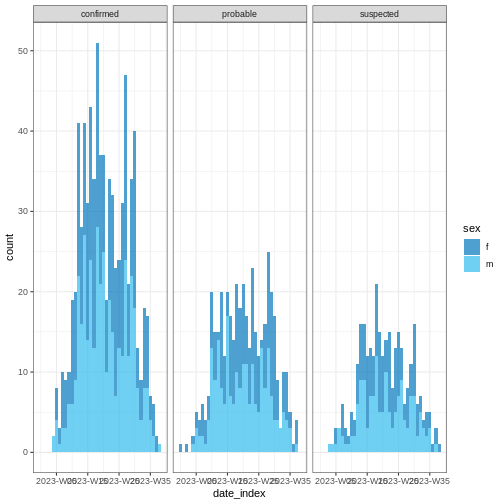
Figure 4
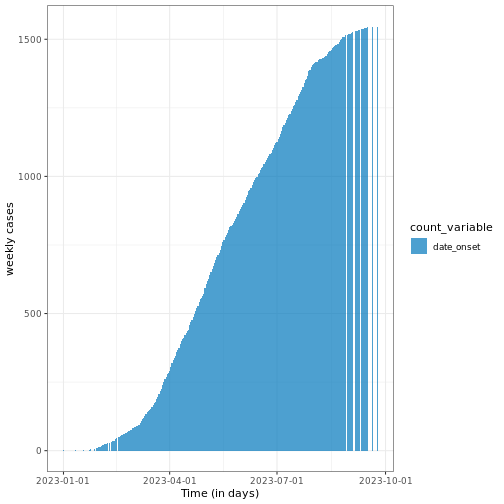
Figure 5
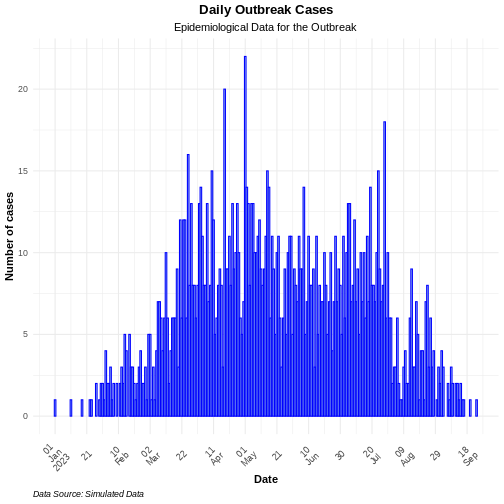
Figure 6
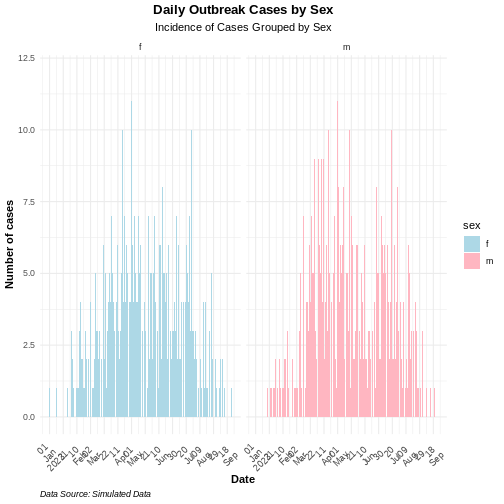
 Pre-Alpha
This lesson is in the pre-alpha phase, which means that it is in early development, but has not yet been taught.
Pre-Alpha
This lesson is in the pre-alpha phase, which means that it is in early development, but has not yet been taught.

Example of data cleaning report generated by cleanepi





