Aggregate and visualize
Last updated on 2026-01-09 | Edit this page
Estimated time: 30 minutes
Overview
Questions
- How to aggregate and summarise case data?
- How to visualize aggregated data?
- What is distribution of cases in time, place, gender, age?
Objectives
- Simulate synthetic outbreak data
- Convert indivdual linelist data to incidence over time
- Create epidemic curves from incidence data
Introduction
In an analytic pipeline, exploratory data analysis (EDA) is an important step before formal modelling. EDA helps determine relationships between variables and summarize their main characteristics, often by means of data visualization.
This episode focuses on EDA of outbreak data using R packages. A key aspect of EDA in epidemic analysis is ‘person, place and time’. It is useful to identify how observed events - such as confirmed cases, hospitalizations, deaths, and recoveries - change over time, and how these vary across different locations and demographic factors, including gender, age, and more.
Let’s start by loading the package incidence2 to
aggregate linelist data according to specific characteristics, and
visualize the resulting epidemic curves (epicurves) that plot the number
of new events (i.e. incidence) over time. We’ll use
simulist to simulate some outbreak data to analyse, and
{tracetheme} for figure formatting. We’ll use the pipe
%>% to connect some of their functions, including others
from the packages dplyr and ggplot2, so
let’s also call to the tidyverse package:
R
# Load packages
library(incidence2) # For aggregating and visualising
library(simulist) # For simulating linelist data
library(tracetheme) # For formatting figures
library(tidyverse) # For {dplyr} and {ggplot2} functions and the pipe %>%
The double-colon
The double-colon :: in R lets you call a specific
function from a package without loading the entire package into the
current environment.
For example, dplyr::filter(data, condition) uses
filter() from the dplyr package. This help
us remember package functions and avoid namespace conflicts.
Synthetic outbreak data
To illustrate the process of conducting EDA on outbreak data, we will generate a line list for a hypothetical disease outbreak utilizing the simulist package. simulist generates simulation data for outbreak according to a given configuration. Its minimal configuration can generate a linelist, as shown in the below code chunk:
R
# Simulate linelist data for an outbreak with size between 1000 and 1500
set.seed(1) # Set seed for reproducibility
sim_data <- simulist::sim_linelist(outbreak_size = c(1000, 1500)) %>%
dplyr::as_tibble() # for a simple data frame output
WARNING
Warning: Number of cases exceeds maximum outbreak size.
Returning data early with 1546 cases and 3059 total contacts (including cases).R
# Display the simulated dataset
sim_data
OUTPUT
# A tibble: 1,546 × 13
id case_name case_type sex age date_onset date_reporting
<int> <chr> <chr> <chr> <int> <date> <date>
1 1 Zahra al-Masri probable f 37 2023-01-01 2023-01-01
2 3 Waleeda al-Muhammad probable f 12 2023-01-11 2023-01-11
3 6 Rhett Jackson confirmed m 53 2023-01-18 2023-01-18
4 8 Sunnique Sims confirmed f 36 2023-01-23 2023-01-23
5 11 Danielle Griggs probable f 77 2023-01-30 2023-01-30
6 14 Mohamed Parker probable m 37 2023-01-24 2023-01-24
7 15 Melissa Eriacho probable f 67 2023-01-31 2023-01-31
8 16 Maria Laughlin probable f 80 2023-01-30 2023-01-30
9 20 Phillip Park confirmed m 70 2023-01-27 2023-01-27
10 21 Dewarren Newton probable m 87 2023-02-09 2023-02-09
# ℹ 1,536 more rows
# ℹ 6 more variables: date_admission <date>, outcome <chr>,
# date_outcome <date>, date_first_contact <date>, date_last_contact <date>,
# ct_value <dbl>This linelist dataset has entries on individual-level simulated events during the outbreak.
The above is the default configuration of simulist, so
includes a number of assumptions about the transmissibility and severity
of the pathogen. If you want to know more about
sim_linelist() and other functionalities check the documentation
website.
You can also find data sets from real emergencies from the past at
the {outbreaks} R
package.
Aggregating
Often we want to analyse and visualise the number of events that
occur on a particular day or week, rather than focusing on individual
cases. This requires grouping linelist data into incidence data. The incidence2
package offers a useful function called
incidence2::incidence() for grouping case data, usually
based around dated events and/or other characteristics. The code chunk
provided below demonstrates the creation of an
<incidence2> class object from the simulated Ebola
linelist data based on the date of onset.
R
# Create an incidence object by aggregating case data based on the date of onset
daily_incidence <- incidence2::incidence(
sim_data,
date_index = "date_onset",
interval = "day" # Aggregate by daily intervals
)
# View the incidence data
daily_incidence
OUTPUT
# incidence: 232 x 3
# count vars: date_onset
date_index count_variable count
<date> <chr> <int>
1 2023-01-01 date_onset 1
2 2023-01-11 date_onset 1
3 2023-01-18 date_onset 1
4 2023-01-23 date_onset 1
5 2023-01-24 date_onset 1
6 2023-01-27 date_onset 2
7 2023-01-29 date_onset 1
8 2023-01-30 date_onset 2
9 2023-01-31 date_onset 2
10 2023-02-01 date_onset 1
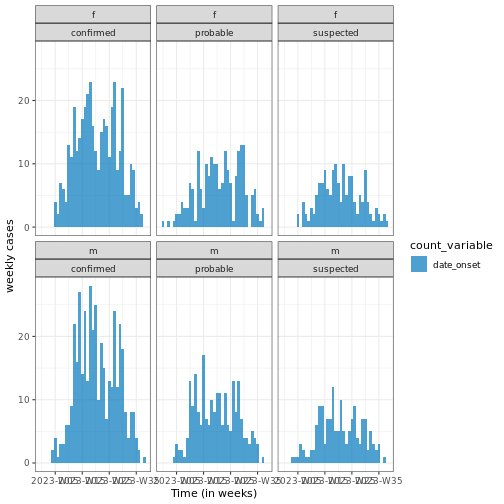
# ℹ 222 more rowsWith the incidence2 package, you can specify the desired interval (e.g. day, week) and categorize cases by one or more factors. Below is a code snippet demonstrating weekly cases grouped by the date of onset, sex, and type of case.
R
# Group incidence data by week, accounting for sex and case type
weekly_incidence <- incidence2::incidence(
sim_data,
date_index = "date_onset",
interval = "week", # Aggregate by weekly intervals
groups = c("sex", "case_type") # Group by sex and case type
)
# View the incidence data
weekly_incidence
OUTPUT
# incidence: 201 x 5
# count vars: date_onset
# groups: sex, case_type
date_index sex case_type count_variable count
<isowk> <chr> <chr> <chr> <int>
1 2022-W52 f probable date_onset 1
2 2023-W02 f probable date_onset 1
3 2023-W03 m confirmed date_onset 1
4 2023-W04 f confirmed date_onset 2
5 2023-W04 f probable date_onset 1
6 2023-W04 m confirmed date_onset 1
7 2023-W04 m probable date_onset 1
8 2023-W05 f confirmed date_onset 3
9 2023-W05 f probable date_onset 4
10 2023-W05 f suspected date_onset 1
# ℹ 191 more rowsDates Completion
When cases are grouped by different factors, it’s possible that the
events involving these groups may have different date ranges in the
resulting incidence2 object. The incidence2
package provides a function called complete_dates() to
ensure that an incidence object has the same range of dates for each
group. By default, missing counts for a particular group will be filled
with 0 for that date.
This functionality is also available as an argument within
incidence2::incidence() adding
complete_dates = TRUE.
R
# Create an incidence object grouped by sex, aggregating daily
daily_incidence_2 <- incidence2::incidence(
sim_data,
date_index = "date_onset",
groups = "sex",
interval = "day", # Aggregate by daily intervals
complete_dates = TRUE # Complete missing dates in the incidence object
)
Challenge 1: Can you do it?
-
Task: Aggregate
sim_datalinelist based on admission date and case outcome in biweekly intervals, and save the results in an object calledbiweekly_incidence.
Visualization
The incidence2 object can be visualized using the
plot() function from the base R package. The resulting
graph is referred to as an epidemic curve, or epi-curve for short. The
following code snippets generate epi-curves for the
daily_incidence and weekly_incidence incidence
objects mentioned above.
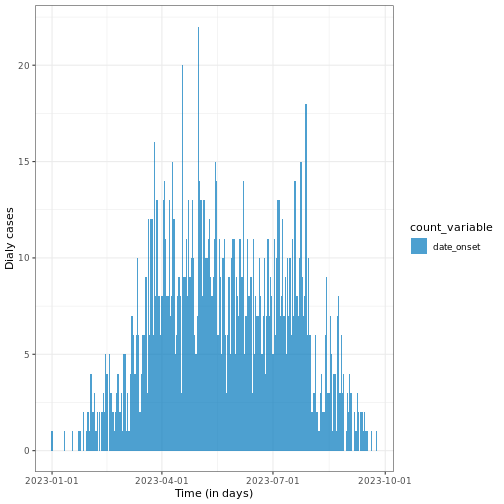
R
# Plot daily incidence data
base::plot(daily_incidence) +
ggplot2::labs(
x = "Time (in days)", # x-axis label
y = "Dialy cases" # y-axis label
) +
tracetheme::theme_trace() # Apply the custom trace theme
R
# Plot weekly incidence data
base::plot(weekly_incidence) +
ggplot2::labs(
x = "Time (in weeks)", # x-axis label
y = "weekly cases" # y-axis label
) +
tracetheme::theme_trace() # Apply the custom trace theme
Easy aesthetics
We invite you to skim the incidence2 package “Get
started” vignette. Find how you can use arguments within
plot() to provide aesthetics to your incidence2 class
objects.
R
base::plot(weekly_incidence, fill = "sex")
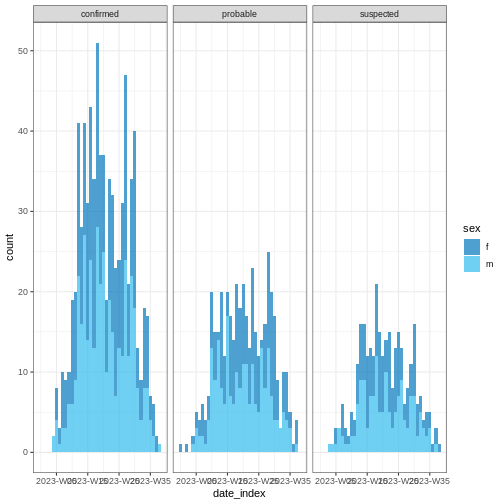
Some of them include show_cases = TRUE,
angle = 45, and n_breaks = 5. Feel free to
give them a try.
Challenge 2: Can you do it?
-
Task: Visualize
biweekly_incidenceobject.
Curve of cumulative cases
The cumulative number of cases can be calculated using the
cumulate() function from an incidence2 object
and visualized, as in the example below.
R
# Calculate cumulative incidence
cum_df <- incidence2::cumulate(daily_incidence)
# Plot cumulative incidence data using ggplot2
base::plot(cum_df) +
ggplot2::labs(
x = "Time (in days)", # x-axis label
y = "weekly cases" # y-axis label
) +
tracetheme::theme_trace() # Apply the custom trace theme
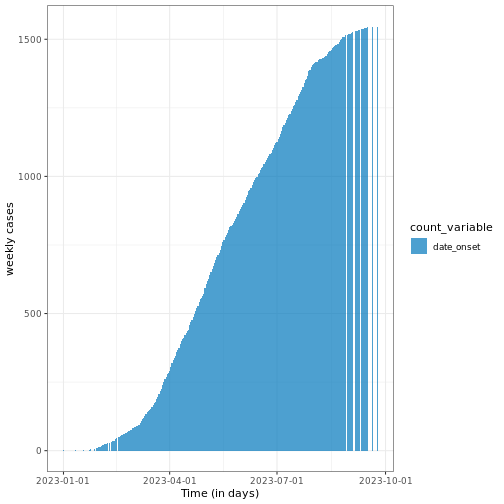
Note that this function preserves grouping, i.e., if the
incidence2 object contains groups, it will accumulate the
cases accordingly.
Challenge 3: Can you do it?
-
Task: Visulaize the cumulatie cases from
biweekly_incidenceobject.
Peak estimation
You can estimate the peak – the time with the highest number of
recorded cases– using the estimate_peak() function from the
{incidence2} package. This function employs a bootstrapping method to
determine the peak time (i.e. by resampling dates with replacement,
resulting in a distribution of estimated peak times).
R
# Estimate the peak of the daily incidence data
peak <- incidence2::estimate_peak(
daily_incidence,
n = 100, # Number of simulations for the peak estimation
alpha = 0.05, # Significance level for the confidence interval
first_only = TRUE, # Return only the first peak found
progress = FALSE # Disable progress messages
)
# Display the estimated peak
print(peak)
OUTPUT
# A tibble: 1 × 7
count_variable observed_peak observed_count bootstrap_peaks lower_ci
<chr> <date> <int> <list> <date>
1 date_onset 2023-05-01 22 <df [100 × 1]> 2023-03-26
# ℹ 2 more variables: median <date>, upper_ci <date>This example demonstrates how to estimate the peak time using the
estimate_peak() function at \(95%\) confidence interval and using 100
bootstrap samples.
Challenge 4: Can you do it?
-
Task: Estimate the peak time from
biweekly_incidenceobject.
Visualization with ggplot2
incidence2 produces basic plots for epicurves, but additional work is required to create well-annotated graphs. However, using the ggplot2 package, you can generate more sophisticated and epicurves with more flexibility in annotation. ggplot2 is a comprehensive package with many functionalities. However, we will focus on three key elements for producing epicurves: histogram plots, scaling date axes and their labels, and general plot theme annotation. The example below demonstrates how to configure these three elements for a simple incidence2 object.
R
# Define date breaks for the x-axis
breaks <- seq.Date(
from = min(as.Date(daily_incidence$date_index, na.rm = TRUE)),
to = max(as.Date(daily_incidence$date_index, na.rm = TRUE)),
by = 20 # every 20 days
)
# Create the plot
ggplot2::ggplot(data = daily_incidence) +
geom_histogram(
mapping = aes(
x = as.Date(date_index),
y = count
),
stat = "identity",
color = "blue", # bar border color
fill = "lightblue", # bar fill color
width = 1 # bar width
) +
theme_minimal() + # apply a minimal theme for clean visuals
theme(
plot.title = element_text(face = "bold",
hjust = 0.5), # center and bold title
plot.subtitle = element_text(hjust = 0.5), # center subtitle
plot.caption = element_text(face = "italic",
hjust = 0), # italicized caption
axis.title = element_text(face = "bold"), # bold axis titles
axis.text.x = element_text(angle = 45, vjust = 0.5) # rotated x-axis text
) +
labs(
x = "Date", # x-axis label
y = "Number of cases", # y-axis label
title = "Daily Outbreak Cases", # plot title
subtitle = "Epidemiological Data for the Outbreak", # plot subtitle
caption = "Data Source: Simulated Data" # plot caption
) +
scale_x_date(
breaks = breaks, # set custom breaks on the x-axis
labels = scales::label_date_short() # shortened date labels
)
WARNING
Warning in geom_histogram(mapping = aes(x = as.Date(date_index), y = count), :
Ignoring unknown parameters: `binwidth` and `bins`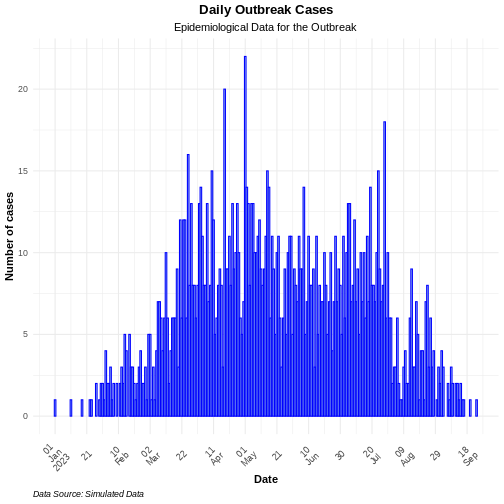
Use the group option in the mapping function to
visualize an epicurve with different groups. If there is more than one
grouping factor, use the facet_wrap() option, as
demonstrated in the example below:
R
# Plot daily incidence by sex with facets
ggplot2::ggplot(data = daily_incidence_2) +
geom_histogram(
mapping = aes(
x = as.Date(date_index),
y = count,
group = sex,
fill = sex
),
stat = "identity"
) +
theme_minimal() + # apply minimal theme
theme(
plot.title = element_text(face = "bold",
hjust = 0.5), # bold and center the title
plot.subtitle = element_text(hjust = 0.5), # center the subtitle
plot.caption = element_text(face = "italic", hjust = 0), # italic caption
axis.title = element_text(face = "bold"), # bold axis labels
axis.text.x = element_text(angle = 45,
vjust = 0.5) # rotate x-axis text for readability
) +
labs(
x = "Date", # x-axis label
y = "Number of cases", # y-axis label
title = "Daily Outbreak Cases by Sex", # plot title
subtitle = "Incidence of Cases Grouped by Sex", # plot subtitle
caption = "Data Source: Simulated Data" # caption for additional context
) +
facet_wrap(~sex) + # create separate panels by sex
scale_x_date(
breaks = breaks, # set custom date breaks
labels = scales::label_date_short() # short date format for x-axis labels
) +
scale_fill_manual(values = c("lightblue",
"lightpink")) # custom fill colors for sex
WARNING
Warning in geom_histogram(mapping = aes(x = as.Date(date_index), y = count, :
Ignoring unknown parameters: `binwidth` and `bins`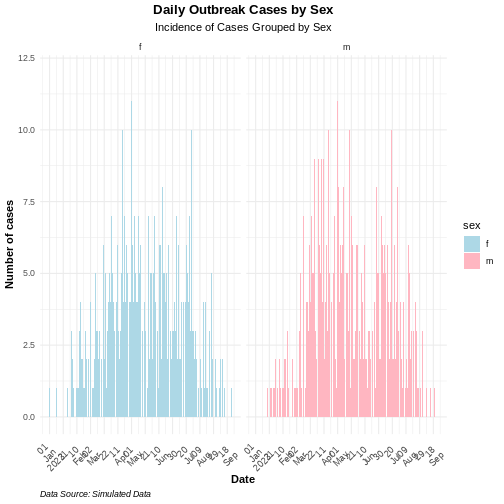
Challenge 5: Can you do it?
- Task: Produce an annotated figure for biweekly_incidence using ggplot2 package.
Key Points
- Use simulist package to generate synthetic outbreak data
- Use incidence2 package to aggregate case data based on a date event, and produce epidemic curves.
- Use ggplot2 package to produce better annotated epicurves.
