Quantifying transmission
Last updated on 2026-02-11 | Edit this page
Estimated time: 30 minutes
Overview
Questions
- How can I estimate the time-varying reproduction number (\(Rt\)) and growth rate from a time series of case data?
- How can I quantify geographical heterogeneity from these transmission metrics?
Objectives
- Learn how to estimate transmission metrics from a time series of
case data using the R package
EpiNow2
Prerequisites
Learners should familiarise themselves with following concepts before working through this tutorial:
Statistics: probability distributions, principle of Bayesian analysis.
Epidemic theory: Effective reproduction number.
Data science: Data transformation and visualization. You can review the episode on Aggregate and visualize incidence data.
Reminder: the Effective Reproduction Number, \(R_t\)
The basic reproduction number, \(R_0\), is the average number of cases caused by one infectious individual in an entirely susceptible population.
But in an ongoing outbreak, the population does not remain entirely susceptible as those that recover from infection are typically immune. Moreover, there can be changes in behaviour or other factors that affect transmission. When we are interested in monitoring changes in transmission we are therefore more interested in the value of the effective reproduction number, \(R_t\), which represents the average number of cases caused by one infectious individual in the population at time \(t\), given the current state of the population (including immunity levels and control measures).
Introduction
The transmission intensity of an outbreak is quantified using two key metrics: the reproduction number, which informs on the strength of the transmission by indicating how many new cases are expected from each existing case; and the growth rate, which informs on the speed of the transmission by indicating how rapidly the outbreak is spreading or declining (doubling/halving time) within a population. For more details on the distinction between speed and strength of transmission and implications for control, review Dushoff & Park, 2021.
To estimate these key metrics using case data we must account for delays between the date of infections and date of reported cases. In an outbreak situation, data are usually available on reported dates only, therefore we must use estimation methods to account for these delays when trying to understand changes in transmission over time.
In the next tutorials we will focus on how to use the functions in EpiNow2 to estimate transmission metrics of case data. We will not cover the theoretical background of the models or inference framework, for details on these concepts see the vignette.
In this tutorial we are going to learn how to use the
EpiNow2 package to estimate the time-varying reproduction
number. We’ll get input data from incidence2. We’ll use
the tidyr and dplyr packages to arrange
some of its outputs, ggplot2 to visualize case
distribution, and the pipe %>% to connect some of their
functions, so let’s also call to the tidyverse
package:
R
library(EpiNow2)
library(incidence2)
library(tidyverse)
The double-colon
The double-colon :: in R lets you call a specific
function from a package without loading the entire package into the
current environment.
For example, dplyr::filter(data, condition) uses
filter() from the dplyr package.
This helps us remember package functions and avoid namespace conflicts.
This tutorial illustrates the usage of epinow() to
estimate the time-varying reproduction number and infection times.
Learners should understand the necessary inputs to the model and the
limitations of the model output.
Bayesian inference
The R package EpiNow2 uses a Bayesian inference framework to
estimate reproduction numbers and infection times based on reporting
dates. In other words, it estimates transmission based on when people
were actually infected (rather than symptom onset), by accounting for
delays in observed data. In contrast, the EpiEstim
package allows faster and simpler real-time estimation of the
reproduction number using only case data over time, reflecting how
transmission changes based on when symptoms appear.
In Bayesian inference, we use prior knowledge (prior distributions) with data (in a likelihood function) to find the posterior probability:
\(Posterior \, probability \propto likelihood \times prior \, probability\)
Refer to the prior probability distribution and the posterior probability distribution.
In the “Expected change in reports”
callout, by “the posterior probability that \(R_t < 1\)”, we refer specifically to the
area
under the posterior probability distribution curve.
Delay distributions and case data
Case data
To illustrate the functions of EpiNow2 we will use
outbreak data of the start of the COVID-19 pandemic from the United
Kingdom, but only with the first 90 days observed. The data are
available in the R package incidence2.
R
dplyr::as_tibble(incidence2::covidregionaldataUK)
OUTPUT
# A tibble: 6,370 × 13
date region region_code cases_new cases_total deaths_new deaths_total
<date> <chr> <chr> <dbl> <dbl> <dbl> <dbl>
1 2020-01-30 East Mi… E12000004 NA NA NA NA
2 2020-01-30 East of… E12000006 NA NA NA NA
3 2020-01-30 England E92000001 2 2 NA NA
4 2020-01-30 London E12000007 NA NA NA NA
5 2020-01-30 North E… E12000001 NA NA NA NA
6 2020-01-30 North W… E12000002 NA NA NA NA
7 2020-01-30 Norther… N92000002 NA NA NA NA
8 2020-01-30 Scotland S92000003 NA NA NA NA
9 2020-01-30 South E… E12000008 NA NA NA NA
10 2020-01-30 South W… E12000009 NA NA NA NA
# ℹ 6,360 more rows
# ℹ 6 more variables: recovered_new <dbl>, recovered_total <dbl>,
# hosp_new <dbl>, hosp_total <dbl>, tested_new <dbl>, tested_total <dbl>To use the data, we must format the data to have two columns:
-
date: the date (as a date object see?is.Date()), -
confirm: number of disease reports (confirm) on that date.
Let’s use tidyr and incidence2 for this:
R
cases_sliced <- incidence2::covidregionaldataUK %>%
tibble::as_tibble() %>%
# Preprocess missing values
tidyr::replace_na(base::list(cases_new = 0)) %>%
# Compute the daily incidence
incidence2::incidence(
date_index = "date",
counts = "cases_new",
count_values_to = "confirm",
date_names_to = "date",
complete_dates = TRUE
) %>%
# Keep the first 90 dates
dplyr::slice_head(n = 90)
With incidence2::incidence() we aggregate cases in
different time intervals (i.e., days, weeks or months) or per
group categories. Also we can have complete dates for all the
range of dates per group category using
complete_dates = TRUE Explore later the incidence2::incidence()
reference manual
Can we replicate {incidence2} with {dplyr}?
We can get an object similar to cases from the
incidence2::covidregionaldataUK data frame using the
dplyr package.
R
incidence2::covidregionaldataUK %>%
dplyr::select(date, cases_new) %>%
dplyr::group_by(date) %>%
dplyr::summarise(confirm = sum(cases_new, na.rm = TRUE)) %>%
dplyr::ungroup() %>%
dplyr::slice_head(n = 90)
However, the incidence2::incidence() function contains
convenient arguments like complete_dates that facilitate
getting an incidence object with the same range of dates for each
grouping without the need of extra code lines or a time-series
package.
In an outbreak situation it is likely we would only have access to the beginning of the input data set. Therefore we assume we only have the first 90 days of this data.
R
plot(cases_sliced)
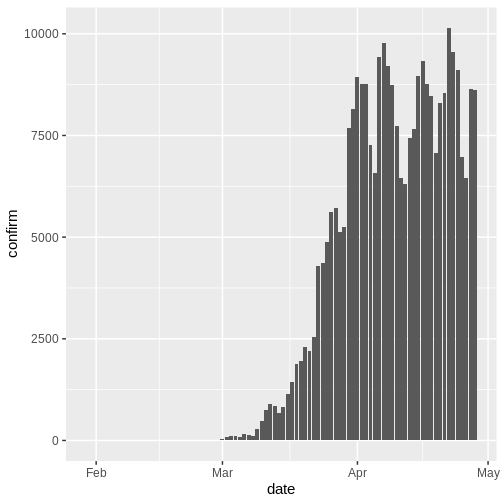
To pass the outputs from incidence2 to
EpiNow2 we need to drop one column from the
cases_sliced object:
R
# Drop column for {EpiNow2} input format
cases <- cases_sliced %>%
dplyr::select(-count_variable)
cases
OUTPUT
# A tibble: 90 × 2
date confirm
<date> <dbl>
1 2020-01-30 3
2 2020-01-31 0
3 2020-02-01 0
4 2020-02-02 0
5 2020-02-03 0
6 2020-02-04 0
7 2020-02-05 2
8 2020-02-06 0
9 2020-02-07 0
10 2020-02-08 8
# ℹ 80 more rowsDelay distributions
We assume there are delays from the time of infection until the time a case is reported. We specify these delays as distributions to account for the uncertainty in individual level differences. The delay may involve multiple types of processes. A typical delay from time of infection to case reporting may consist of:
time from infection to symptom onset (the incubation period) + time from symptom onset to case notification (the reporting time) .
The delay distribution for each of these processes can either estimated from data or obtained from the literature. We can express uncertainty about what the correct parameters of the distributions by assuming the distributions have fixed parameters or whether they have variable parameters. To understand the difference between fixed and variable distributions, let’s consider the incubation period.
Delays and data
The number of delays and type of delay are a flexible input that depend on the data. The examples below highlight how the delays can be specified for different data sources:
| Data source | Delay(s) |
|---|---|
| Time of symptom onset | Incubation period |
| Time of case report | Incubation period + time from symptom onset to case notification |
| Time of hospitalisation | Incubation period + time from symptom onset to hospitalisation |
Incubation period distribution
The distribution of incubation period for many diseases can usually be obtained from the literature. The package epiparameter contains a library of epidemiological parameters for different diseases obtained from the literature.
We specify a (fixed) gamma distribution with mean \(\mu = 4\) and standard deviation \(\sigma = 2\) (shape = \(4\), scale = \(1\)) using the function
Gamma() as follows:
R
incubation_period_fixed <- EpiNow2::Gamma(
mean = 4,
sd = 2,
max = 20
)
incubation_period_fixed
OUTPUT
- gamma distribution (max: 20):
shape:
4
rate:
1The argument max is the maximum value the distribution
can take; in this example, 20 days.
We can plot distributions generated by EpiNow2 using
plot():
R
plot(incubation_period_fixed)
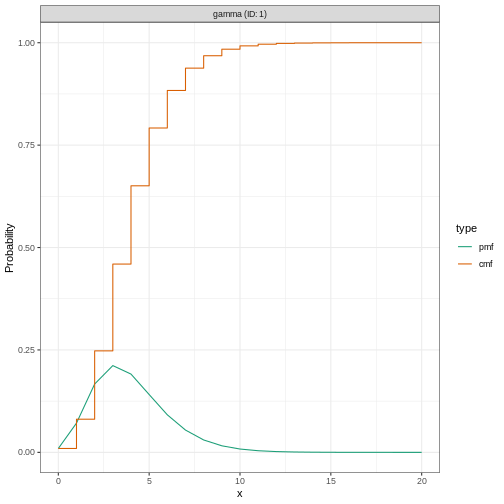
Why a gamma distribution?
The incubation period must be a positive value. Therefore we must specify a distribution in EpiNow2 which is for positive values only.
Gamma() supports Gamma distributions and
LogNormal() Log-normal distributions, which are
distributions for positive values only.
For all types of delay, we will need to use distributions for positive values only - we don’t want to include delays of negative days in our analysis!
Including distribution uncertainty
To specify a variable distribution, we include uncertainty around the mean \(\mu\) and standard deviation \(\sigma\) of our gamma distribution. If our incubation period distribution has a mean \(\mu\) and standard deviation \(\sigma\), then we assume the mean (\(\mu\)) follows a Normal distribution with standard deviation \(\sigma_{\mu}\):
\[\mbox{Normal}(\mu,\sigma_{\mu}^2)\]
and a standard deviation (\(\sigma\)) follows a Normal distribution with standard deviation \(\sigma_{\sigma}\):
\[\mbox{Normal}(\sigma,\sigma_{\sigma}^2).\]
We specify this using Normal() for each argument: the
mean (\(\mu = 4\) with \(\sigma_{\mu} = 0.5\)) and standard
deviation (\(\sigma = 2\) with \(\sigma_{\sigma} = 0.5\)).
R
incubation_period_variable <- EpiNow2::Gamma(
mean = EpiNow2::Normal(mean = 4, sd = 0.5),
sd = EpiNow2::Normal(mean = 2, sd = 0.5),
max = 20
)
incubation_period_variable
OUTPUT
- gamma distribution (max: 20):
shape:
- normal distribution:
mean:
4
sd:
0.61
rate:
- normal distribution:
mean:
1
sd:
0.31Let’s plot the distribution we just configured:
R
plot(incubation_period_variable)
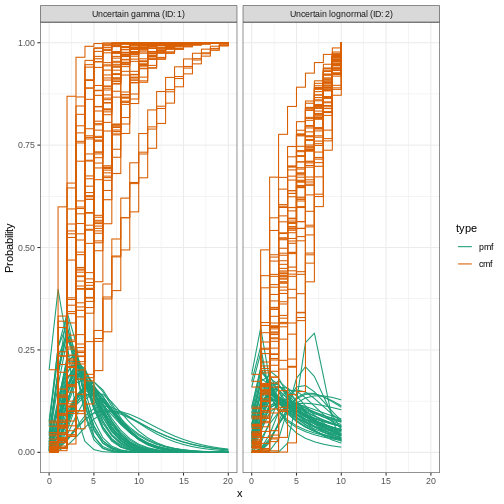
Reporting delays
After the incubation period, there will be an additional delay of time from symptom onset to case notification: the reporting delay. We can specify this as a fixed or variable distribution, or estimate a distribution from data.
When specifying a distribution, it is useful to visualise the probability density to see the peak and spread of the distribution, in this case we will use a log normal distribution.
If we want to assume that the mean reporting delay is 2 days (with a
uncertainty of 0.5 days) and a standard deviation of 1 day (with
uncertainty of 0.5 days), we can specify a variable distribution using
LogNormal() as before:
R
reporting_delay_variable <- EpiNow2::LogNormal(
meanlog = EpiNow2::Normal(mean = 2, sd = 0.5),
sdlog = EpiNow2::Normal(mean = 1, sd = 0.5),
max = 10
)
Visualize a log Normal distribution using {epiparameter}
Using epiparameter::epiparameter() we can create a
custom distribution. The fixed log normal distribution will look
like:
R
library(epiparameter)
R
epiparameter::epiparameter(
disease = "covid",
epi_name = "reporting delay",
prob_distribution =
epiparameter::create_prob_distribution(
prob_distribution = "lnorm",
prob_distribution_params = c(
meanlog = 2,
sdlog = 1
)
)
) %>%
plot()
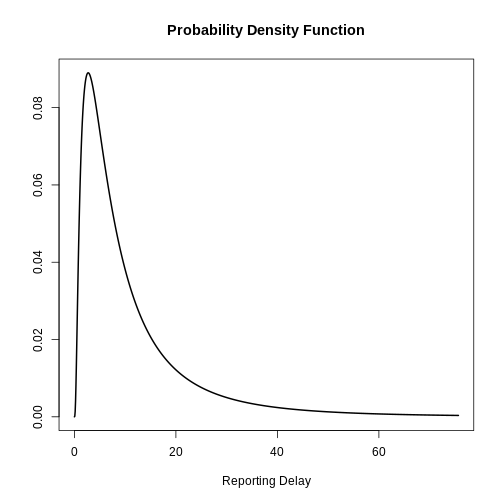
How to get the reporting delay from data?
If data is available on the time between symptom onset and reporting,
we can use the function EpiNow2::estimate_delay() to
estimate a log normal distribution from a vector of delays.
The code below illustrates how to use
EpiNow2::estimate_delay() with synthetic Ebola data. We
calculate the reporting delay for each case the time difference
from date of onset to date of
hospitalization (date when case was reported).
R
library(tidyverse)
# Steps:
# - get Ebola data from package {outbreaks}
# - keep a subset of columns for this example only
# - calculate the time difference between two dates in linelist
# - extract the time difference as a vector class object
# - estimate the delay parameters using {EpiNow2}
outbreaks::ebola_sim_clean$linelist %>%
tibble::as_tibble() %>%
dplyr::select(case_id, date_of_onset, date_of_hospitalisation) %>%
dplyr::mutate(reporting_delay = date_of_hospitalisation - date_of_onset) %>%
dplyr::pull(reporting_delay) %>%
EpiNow2::estimate_delay(
samples = 1000,
bootstraps = 10
)
OUTPUT
- lognormal distribution (max: 22):
meanlog:
- normal distribution:
mean:
0.18
sd:
0.14
sdlog:
- normal distribution:
mean:
1
sd:
0.11Generation time
We also must specify a distribution for the generation time. Here we will use a log normal distribution with mean 3.6 and standard deviation 3.1 (Ganyani et al. 2020).
R
generation_time_variable <- EpiNow2::LogNormal(
mean = EpiNow2::Normal(mean = 3.6, sd = 0.5),
sd = EpiNow2::Normal(mean = 3.1, sd = 0.5),
max = 20
)
Finding estimates
The function epinow() is a wrapper for the function
estimate_infections() used to estimate cases by date of
infection. The generation time distribution and delay distributions must
be passed using the functions generation_time_opts() and
delay_opts() respectively.
There are numerous other inputs that can be passed to
epinow(), see ?EpiNow2::epinow() for more
detail. One optional input is to specify a log normal prior for
the effective reproduction number \(R_t\) at the start of the outbreak. We
specify a mean of 2 days and standard deviation of 2 days as arguments
of prior within rt_opts():
R
# define Rt prior distribution
rt_prior <- EpiNow2::rt_opts(prior = EpiNow2::LogNormal(mean = 2, sd = 2))
Bayesian inference using Stan
The Bayesian inference is performed using MCMC methods with the
program Stan. There are a number of
default inputs to the Stan functions including the number of chains and
number of samples per chain (see
?EpiNow2::stan_opts()).
To reduce computation time, we can run chains in parallel. To do
this, we must set the number of cores to be used. By default, 4 MCMC
chains are run (see stan_opts()$chains), so we can set an
equal number of cores to be used in parallel as follows:
R
withr::local_options(base::list(mc.cores = 4))
To find the maximum number of available cores on your machine, use
parallel::detectCores().
Checklist
Note: In the code below _fixed
distributions are used instead of _variable (delay
distributions with uncertainty). This is to speed up computation time.
It is generally recommended to use variable distributions that account
for additional uncertainty.
R
# fixed alternatives
generation_time_fixed <- EpiNow2::LogNormal(
mean = 3.6,
sd = 3.1,
max = 20
)
reporting_delay_fixed <- EpiNow2::LogNormal(
mean = 2,
sd = 1,
max = 10
)
Now you are ready to run EpiNow2::epinow() to estimate
the time-varying reproduction number for the first 90 days:
R
estimates <- EpiNow2::epinow(
# reported cases
data = cases,
# delays
generation_time = EpiNow2::generation_time_opts(generation_time_fixed),
delays = EpiNow2::delay_opts(incubation_period_fixed + reporting_delay_fixed),
# prior
rt = rt_prior
)
Do not wait for this to continue
For the purpose of this tutorial, we can optionally use
EpiNow2::stan_opts() to reduce computation time. We can
specify a fixed number of samples = 1000 and
chains = 2 to the stan argument of the
EpiNow2::epinow() function. We expect this to take
approximately 3 minutes.
R
# you can add the `stan` argument
EpiNow2::epinow(
...,
stan = EpiNow2::stan_opts(samples = 1000, chains = 3)
)
Remember: Using an appropriate number of samples and chains is crucial for ensuring convergence and obtaining reliable estimates in Bayesian computations using Stan. More accurate outputs come at the cost of speed.
Results
We can extract and visualise estimates of the effective reproduction number through time:
R
estimates$plots$R
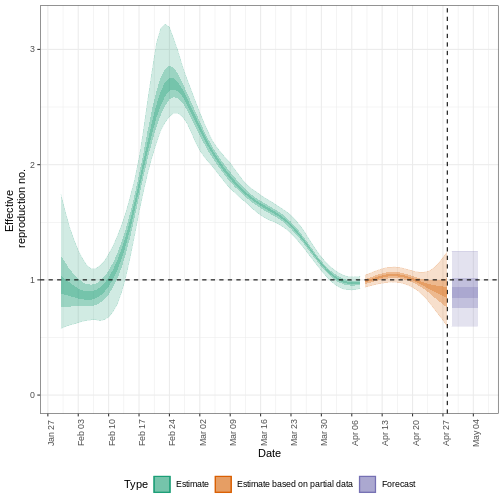
The uncertainty in the estimates increases through time. This is because estimates are informed by data in the past - within the delay periods. This difference in uncertainty is categorised into Estimate (green) utilises all data and Estimate based on partial data (orange) estimates that are based on less data (because infections that happened at the time are more likely to not have been observed yet) and therefore have increasingly wider intervals towards the date of the last data point. Finally, the Forecast (purple) is a projection ahead of time.
We can also visualise the growth rate estimate through time:
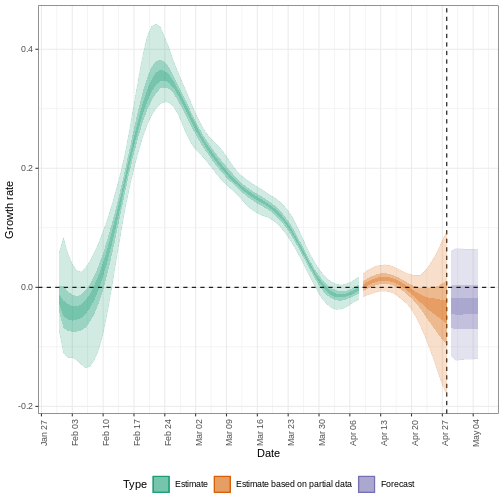
R
estimates$plots$growth_rate
To extract a summary of the key transmission metrics at the latest date in the data:
R
summary(estimates)
OUTPUT
measure estimate
<char> <char>
1: New infections per day 7904 (4778 -- 13261)
2: Expected change in reports Likely decreasing
3: Effective reproduction no. 0.96 (0.74 -- 1.3)
4: Rate of growth -0.013 (-0.1 -- 0.084)
5: Doubling/halving time (days) -55 (8.2 -- -6.6)As these estimates are based on partial data, they have a wide uncertainty interval.
From the summary of our analysis we see that the expected change in reports is Likely decreasing with the estimated new infections 7904 (4778 – 13261).
The effective reproduction number \(R_t\) estimate (on the last date of the data) is 0.96 (0.74 – 1.3).
The exponential growth rate of case numbers is -0.013 (-0.1 – 0.084).
The doubling time (the time taken for case numbers to double) is -55 (8.2 – -6.6).
Expected change in reports
A factor describing the expected change in reports based on the posterior probability that \(R_t < 1\).
| Probability (\(p\)) | Expected change |
|---|---|
| \(p < 0.05\) | Increasing |
| \(0.05 \leq p< 0.4\) | Likely increasing |
| \(0.4 \leq p< 0.6\) | Stable |
| \(0.6 \leq p < 0.95\) | Likely decreasing |
| \(0.95 \leq p \leq 1\) | Decreasing |
Credible intervals
In all EpiNow2 output figures, shaded regions reflect 90%, 50%, and 20% credible intervals in order from lightest to darkest.
Checklist
EpiNow2 can be used to estimate transmission metrics
from case data at any time in the course of an outbreak. The reliability
of these estimates depends on the quality of the data and appropriate
choice of delay distributions. In the next tutorial we will learn how to
make forecasts and investigate some of the additional inference options
available in EpiNow2.
How does {EpiNow2} work?
{EpiNow2} contains different models for how infections arise. One of the main models that is used in the default configuration is the Renewal equation model. If you want to gain familiarity with it and learn how delays are accounted for when estimating \(R_t\), you can read the episode on Introduction to the Renewal equation in the “More Resources” section of this tutorial’s website.
Challenge
Challenge
Quantify geographical heterogeneity
The outbreak data of the start of the COVID-19 pandemic from the United Kingdom from the R package incidence2 includes the region in which the cases were recorded. To find regional estimates of the effective reproduction number and cases, we must format the data to have three columns:
-
date: the date, -
region: the region, -
confirm: number of disease reports (confirm) for a region on a given date.
Generate regional Rt estimates from the
incidence2::covidregionaldataUK data frame by:
- use incidence2 to convert aggregated data to
incidence data by the variable
region, - keep the first 90 dates for all regions,
- estimate the Rt per region using the defined generation time and delays in this episode.
R
regional_cases <- incidence2::covidregionaldataUK %>%
# use {tidyr} to preprocess missing values
tidyr::replace_na(base::list(cases_new = 0))
To wrangle data, you can:
R
regional_cases <- incidence2::covidregionaldataUK %>%
# use {tidyr} to preprocess missing values
tidyr::replace_na(base::list(cases_new = 0)) %>%
# use {incidence2} to convert aggregated data to incidence data
incidence2::incidence(
date_index = "date",
groups = "region",
counts = "cases_new",
count_values_to = "confirm",
date_names_to = "date",
complete_dates = TRUE
) %>%
dplyr::select(-count_variable) %>%
dplyr::filter(date < ymd(20200301))
To learn how to do the regional estimation of Rt, read the Get
started vignette section on regional_epinow() at https://epiforecasts.io/EpiNow2/articles/EpiNow2.html#regional_epinow
To find regional estimates, we use the same inputs as
epinow() to the function
regional_epinow():
R
estimates_regional <- EpiNow2::regional_epinow(
# cases
data = regional_cases,
# delays
generation_time = EpiNow2::generation_time_opts(generation_time_fixed),
delays = EpiNow2::delay_opts(incubation_period_fixed + reporting_delay_fixed),
# prior
rt = rt_prior
)
Plot the results with:
R
estimates_regional$summary$summarised_results$table
estimates_regional$summary$plots$R
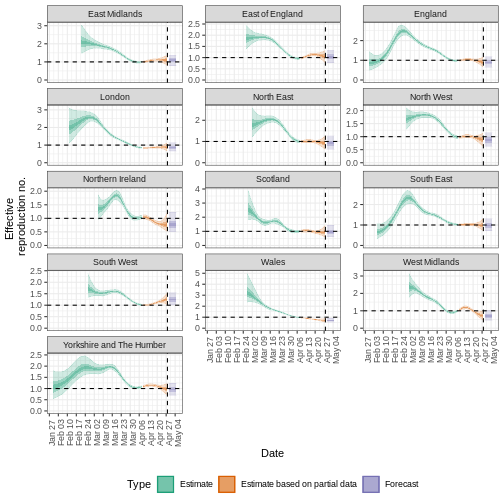
Key Points
- Transmission metrics can be estimated from case data after accounting for delays
- Uncertainty can be accounted for in delay distributions
