This vignette outlines the design decisions that have been taken during the development of the epidemics R package, and provides some of the reasoning, and possible pros and cons of each decision.
This document is primarily intended to be read by those interested in understanding the code within the package and for potential package contributors.
Scope
epidemics aims to help public health practitioners - rather than research-focussed modellers - to rapidly simulate disease outbreak scenarios, and aims to balance flexibility, performance, user-friendliness, and maintainability.
epidemics trades away some flexibility in defining model structures for a gain in the ease of defining epidemic scenario components such as affected populations and model events.
epidemics attempts to balance performance and maintainability through minimum sufficient use of C++, and not attempting to write a domain-specific language (such as odin).
To be more broadly applicable, epidemics provides a ‘library’ of compartmental models which are adapted from the published literature. These models focus on broad types of diseases or settings, rather than specific diseases or scenarios. Thus all models are intended to be applicable to a range of diseases.
Output
All function outputs are expected to return a structure that inherits from <data.frame>, to make further processing easier for users.
The exact structure of the output - whether a list of <data.frame>s, a <data.table> or <tibble>, or a nested version of these tabular data classes - is yet to be fixed.
The eventual stable output type must allow users to conveniently access the epidemic trajectory data, identify and filter intervention scenarios for comparisons among them, and allow interoperability with data science tools such as the Tidyverse.
Package architecture

Fig. 1: epidemics is designed to allow easy combination of composable elements with a model structure taken from a library of published models, with sensible default parameters, to allow public health practitioners to conveniently model epidemic scenarios and the efficacy of response strategies.
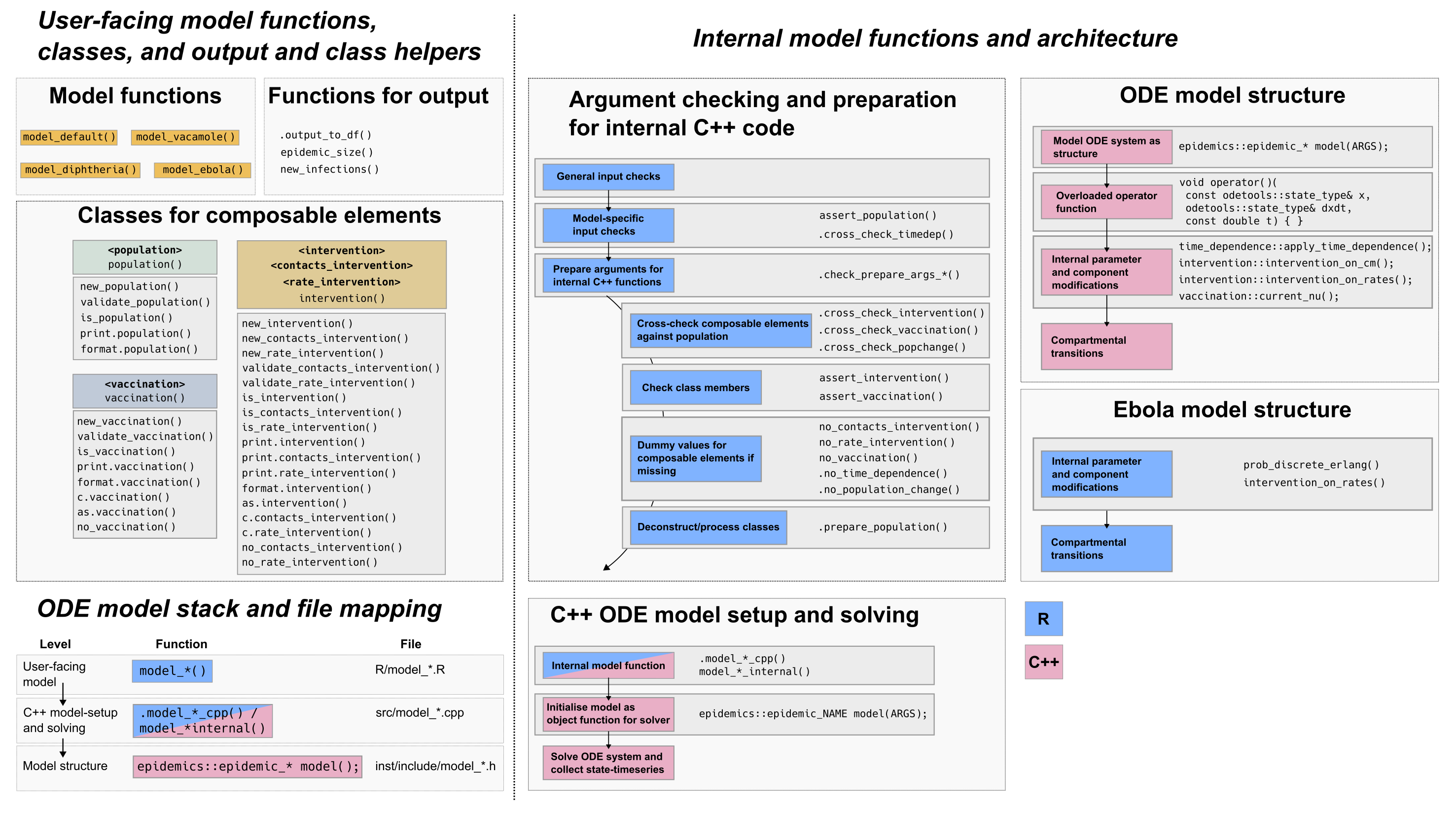
Fig. 2: epidemics package architecture, and the ODE model stack. epidemics includes multiple internal functions in both R and C++ which check that inputs are suitable for each model, and to cross-check that inputs are compatible with each other. Function names indicate their behaviour (e.g. assert_*()), but not all similarly named functions are called at similar places in model function bodies. Partly, this reflects the privileged positions of some model components (such as the population, against which other components are checked), but also that some composable elements (such as time-dependence) may not be vectorised.
Design decisions
Epidemic modelling
The notes here refer to broad decisions and not to individual models – see individual model vignettes and published sources for modelling decisions.
-
There are two broad types of models:
Deterministic models implemented as solutions to systems of ordinary different equations (ODEs),
Stochastic models with discrete timesteps where individuals move between compartments probabilistically.
epidemics models’ compartmental transitions are fixed. Users cannot create new links between compartments, although links between compartments can be disallowed by modifying model parameters.
The models’ epidemiological parameter sets are unique, although some parameters are shared among models. These can be changed by users although reasonable default values are provided.
The model components - such as the population affected by the epidemic, or any policy responses in the form of interventions - are called composable elements. Composable elements can be and are expected to be modified by users to match their situation. All model components except the
populationare optional.Each model allows a unique sets of composable elements considered suitable for the expected use case. E.g. the Ebola model does not allow modelling a vaccination regime, as this is considered unlikely for most Ebola outbreaks. The allowed set of composable elements is subject to change, and is open to user requests.
The effect of interventions is modelled as being additive, not multiplicative. When an intervention with an X% reduction in contacts overlaps with an intervention with a Y% reduction, the cumulative effect on contacts \(C\) is \(C \times 1 - (X + Y)\), rather than \(C \times (1 - X)(1 - Y)\). Additive effects were considered easier for users to understand than multiplicative effects.
ODE systems and models
A common method for defining and solving ODEs in R is to write the ODE system in R and pass it to solvers such as
deSolve::lsoda()from the deSolve package. We have opted against this approach (referred to as ‘R-deSolve’).It is possible to write ODE compartmental transitions in C++, and expose the C++ code to R, eventually passing this ODE system to deSolve solvers (referred to as ‘Rcpp-deSolve’). We have also opted against this approach, as it involves substantial interconversion of more complex objects such as structured lists (the model composable elements) from R to C++ in each solver timestep, reducing performance.
ODE systems are written in C++, and solved using the Boost odeint solvers with fixed step sizes. This means that within each call to
model_*(), R objects are converted for use by C++ only once (in the scalar arguments case), and are not passed back and forth multiple times.odeint is provided by the package BH (Boost Headers), making it convenient to use in Rcpp packages. The r2sundials package provides an Rcpp-friendly interface to the SUNDIALS ODE solvers, but the documentation is less clear overall, and seems to recommend the use of RcppArmadillo and RcppXPtrUtils; these have not been evaluated for use.
odeint imposes certain constraints on ODE systems. There is no functionality to easily define ‘events’ (also called ‘callbacks’) and combine them with the ODE system. This means that the ODE systems have to encapsulate information on the timing of events (such as interventions) and the effect.
Equations describing the right-hand side of model ODE systems \(x' = f(x)\) are thus written as
structswith an overloaded function call operator that makes themFunctionObjecttypes. Thestructthus holds epidemiological parameters and composable elements, which are passed by reference in any operations, reducing copying. See this simple example from the Boost documentation. It can be passed as a function to a solver, and the epidemiological parameters are calculated in each solver timestep as some function of time and the composable elements (as applicable).epidemics relies on the Eigen C++ library provided by RcppEigen to represent matrices of initial conditions. Eigen matrices are among the types accepted as initial states by odeint solvers. Alternatives include Boost Basic Linear Algebra Library (uBLAS) and standard library arrays and vectors; a partial list - which does not include Eigen - is provided by Boost, and Armadillo matrices may also be supported. We use Eigen matrices as Eigen offers broad and well documented support for matrix operations, easy conversion between Rcpp types, and also conforms to previous use of Eigen in finalsize.
The models’ C++ code is in two levels that underlie the R source code - the C++ source code and the C++ headers. This is to make the header code (the model ODEs and helper functions) shareable so that they can be re-used in other Rcpp packages and this is explored more in this blog post.
epidemics defines C++ namespaces in the package headers to more clearly indicate where certain functionalities sit. All model structures are in the namespace
epidemics, while the namespacesvaccination,intervention, andtime_dependencecontain C++ code to handle these composable elements (such as calculating the effect of cumulative interventions). The namespaceodetoolsdefines the initial state type, which is anEigen::MatrixXd.
Stochastic models
epidemics currently includes a single discrete-time stochastic model that implements a sub-compartment system taken from @getz2018, and initially adapted in Epirecipes. Some modelling decisions are explained here, while recognising that adding more models with different implementations could see them being revisited.
Stochastic models currently target, and are expected to target, diseases for which outbreak sizes are initially small and thus where stochasticity matters more; the typical example is Ebola virus disease (simply, Ebola), and this is reflected in the model name.
The Ebola model is also expected to be suitable for diseases with a high fatality risk that are detected when outbreak sizes are small, and with an isolation and treatment response comparable to Ebola, e.g. Marburg virus disease.
Note that very different alternative implementations of stochastic models exist.
The current implementation was chosen due to the use of Erlang sub-compartments that were felt to better represent the long-tailed distributions of infection times seen in haemorrhagic fevers like Ebola.
The model implements a two-level structure similar to the ODE models, with
.model_ebola_internal()the function implementing the simulation, whilemodel_ebola()is the user-facing wrapper with input checks and parameter and scenario combination handling.The model allows vectors for the infection parameters and the duration (called
time_end); however the number of replicates for each parameter-scenario combination is not allowed to vary. A default of 100 was chosen as a reasonable middle ground between efficiency and realism.-
The model allows lists of intervention sets and time-dependence functions to be passed.
Only interventions on model parameters are allowed, and interventions on social contacts are not allowed, as the model does not include age-stratification in transmission (or otherwise).
The parameters
infectiousness_rate(often denoted \(\sigma\)) andremoval_rate(recovery or death with safe burial, \(\gamma\)) cannot be targeted by rate interventions or time-dependence as these values determine the number of sub-compartments in the exposed and infectious (and hospitalised) compartments, respectively. These are fixed at the start of the simulation, and changing them partway is challenging to support as it would involve redistributing individuals currently in any sub-compartment into more or fewer sub-compartments.
-
The model supports and defaults to multiple replicates or runs for each parameter-scenario combination.
For any \(N\) runs with a single parameter set and a single intervention set, runs will differ due to stochasticity (essentially, as different random numbers are drawn for each run).
For any \(N\) runs with multiple parameter sets and a single intervention set, runs will differ due to stochasticity within parameter sets, but any differences between runs across parameter sets (e.g. two different transmission rates) will be due to differences in parameter values alone.
For multiple runs of multiple parameter-scenario combinations, differences among runs will be due to differences in parameters and composable elements (such as interventions) alone.
The withr package is used to ensure that seeds are preserved across parameter sets and interventions.
The input and output types correspond to the ODE model types.
-
The model is written in R although this may change in future. The main reason for keeping the implementation in R is that the
stats::rmultinom()function is a very efficient way of drawing from a categorical distribution (often called a discrete distribution) with heterogeneous probabilities. This StackOverflow question from 2014 has more details including benchmarks against other implementations using Rcpp; though old, we have benchmarked a minimal example using Boost and found that this information is still valid.- One possible reason to switch at least part of the Ebola model implementation to C++ is to make use of more efficient random number generation from the GNU Scientific Library via RcppGSL, although there may be issues for users trying to compile from source on Windows.
Classes
The major composable elements are bundled into custom S3 classes that inherit from lists, and which are expected to be understandable elements of epidemic response:
<population>, the<intervention>superclass, and<vaccination>. All other composable elements take the form of named, structured lists. These are not defined as classes because they are expected to be less used, or used by more advanced modellers who are comfortable working with lists directly.A key element of composable elements being or inheriting from lists is that they can be easily handled within the C++ code as they are interpreted as Rcpp lists.
All matrix objects referring to demography-group coefficients follow the ‘demography-in-rows’ pattern from finalsize, where rows represent demography groups, and columns represent coefficients. This includes the initial conditions matrix in
<population>s, where columns are epidemiological compartments, but also vaccination rates in<vaccination>, and contacts reductions in<contacts_intervention>s.Interventions may be of the
<contacts_intervention>or<rate_intervention>class, which inherit from the abstract super-class<intervention>. This inheritance structure was chosen to maintain coherence between the intervention types, and to keep the option open of unifying these classes into a single concrete type in the future.All composable elements except the
populationare optional. Model functions internally generate dummy values for the other composable elements allowed for each model, as these are required for the C++ code.-
<intervention>and<vaccination>objects may be combined with objects of the same (sub)-class using thec()method for each (sub)-class, and resulting in an object of the same (sub)-class. The interpretation for each class is however slightly different:<intervention>: Combining two interventions of the same sub-class is understood to represent the combined application of multiple epidemic response strategies, e.g. closing schools and also closing workplaces, or introducing mask mandates over different intervals.<vaccination>: Combining two vaccination objects results in a two-dose vaccination regime, rather than two separate vaccination campaigns (each delivering a single dose). This is because epidemics focuses on the initial pandemic response phase where vaccination is not expected to be available at scale, rather than long-term vaccination campaigns against endemic infections.
Function vectorisation
epidemics is moving towards allowing vectors or lists to be passed as model function arguments, to easily allow the incorporation of parameter uncertainty and comparisons of multiple scenarios (while maintaining comparability across scenarios in each function call). Consequently, each function call may consist of many 1000s of model runs (see PR #176 and the vignette on scenario modelling).
All model epidemiological parameters are allowed to be passed as numeric vectors (currently only ODE models). This includes
time_end, which allows runs of different durations within a single function call. This use case was taken from this report which aimed to calculate the epidemic size over a given period with uncertainty in the start time (and hence the duration). epidemics follows the Tidyverse rules on vector recycling for epidemiological parameters.Arguments
interventionandvaccinationaccepting composable elements are allowed to be passed as lists of inputs. In the case ofintervention, this is a list of lists, with each element a list of<intervention>objects (typically one<contacts_intervention>and a variable number of<rate_intervention>s): this is referred to as an intervention set. All other composable elements may currently only be scalar values, but this may change in future (see issue #181 for passing lists of populations).Model functions internally create combinations of composable elements, and each such combination is referred to as a scenario, and also combine each scenario with each set of epidemiological parameters. This is to ensure comparability across scenarios.
Miscellaneous decisions
epidemics follows the finalsize example in following Google’s C++ code style, and in using Cpplint and Cppcheck for static code analysis as options such as Clang-tidy do not work well with Rcpp code; see this blog post for more.
Function naming: Function names aim to contain verbs that indicate the function behaviour. Internal input checking functions follow the checkmate naming style (e.g.
assert_*()).Function naming: Internal functions aim to begin with a
.(e.g..model_default_cpp()) to more clearly indicate that they are not for direct use.
Dependencies
The aim is to restrict the number of hard dependencies while maintaining user-friendliness.
- checkmate: a useful input-checking package used across Epiverse packages;
- cli and glue: convenience packages for pretty printing methods; these could be reconsidered to lighten package dependencies;
- data.table: a lightweight dependency to handle data from model outputs, and especially used for nested list column functionality.
- Rcpp: provides linking between C++ and R;
- RcppEigen: provides matrix classes suitable for use with Boost odeint;
- BH: provides the Boost odeint ODE solvers;
- withr: for seed management;
- stats and utils: already installed with R, and used only in the Ebola model, and hence liable to be removed in the future.
A wider range of packages are taken on as soft dependencies to make the vignettes more user-friendly.
- bench: to benchmark epidemics against finalsize;
- bookdown, knitr, rmarkdown: packages to generate vignettes and format them;
- dplyr, tibble, tidyr: to show the processing of model outputs;
- EpiEstim: to show links with upstream packages that estimate R in a vignette;
- finalsize: for comparison against epidemics in a vignette;
- ggplot2, scales, colorspace: packages for plotting;
- socialmixr: to obtain social contacts matrices;
- spelling: for spell-checking;
- testthat (>= 3.0.0): for unit tests.
Contribute
There are no special requirements to contributing to epidemics, but contributions of new models should be clearly motivated, fall within the scope of the package, target a disease type or situation that is not already covered by existing models, and ideally should already be peer-reviewed. In general, please follow the package contributing guide.